6BR1
 
 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 4a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)pyrido[2,3-d]pyrimidine, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
6JMF
 
 | |
6BW8
 
 | | Mcl-1 complexed with small molecules | | Descriptor: | 7-{8-chloro-11-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-oxo-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-4,5-dihydro-1H-[1,4]diazepino[1,2-a]indol-2(3H)-yl}-1-methyl-1H-indole-3-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2017-12-14 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Optimization of Potent and Selective Tricyclic Indole Diazepinone Myeloid Cell Leukemia-1 Inhibitors Using Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
6XSW
 
 | | Structure of the Notch3 NRR in complex with an antibody Fab Fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-N3 Fab Heavy Chain, ... | | Authors: | Bard, J. | | Deposit date: | 2020-07-16 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | NOTCH3-targeted antibody drug conjugates regress tumors by inducing apoptosis in receptor cells and through transendocytosis into ligand cells.
Cell Rep Med, 2, 2021
|
|
8ED6
 
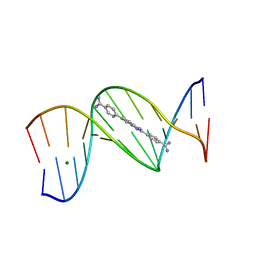 | |
8EDA
 
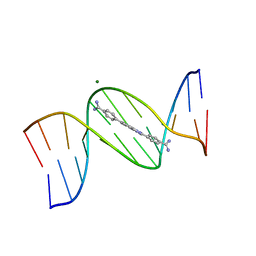 | | An alternating AT dodecamer benzimidazole (DB1476) complex | | Descriptor: | 4,4'-(1H-benzimidazole-2,6-diyl)di(benzene-1-carboximidamide), DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-09-03 | | Release date: | 2023-02-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
8F1S
 
 | | A benzimidazole (DB1476) sequence-specific recognition of 5'-CGCAAAAAAGCG-3' in A-orientation | | Descriptor: | 4,4'-(1H-benzimidazole-2,6-diyl)di(benzene-1-carboximidamide), DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*TP*GP*CP*G)-3'), ... | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-11-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
8F2W
 
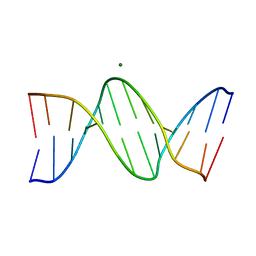 | | Structure of a B-Form Dodecamer: 5'-CGCGAATTCGCG-3 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-11-08 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
8F1V
 
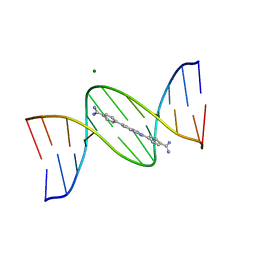 | | A benzimidazole (DB1476) sequence-specific recognition of 5'-CGCAAAAAAGCG-3' in B-orientation | | Descriptor: | 4,4'-(1H-benzimidazole-2,6-diyl)di(benzene-1-carboximidamide), DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*TP*GP*CP*G)-3'), ... | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-11-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
8F2Y
 
 | |
8EDB
 
 | | 5'-CGCGAATTCGCG-3' and an AT-specific binder (DB1884) complex | | Descriptor: | 4,4'-(1H-indole-2,6-diyl)dibenzenecarboximidamide, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-09-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
8F20
 
 | | Structure of an A-tract B-DNA Dodecamer: 5'-CGCAAAAAAGCG-3 | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-11-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
6CE8
 
 | | Crystal structure of fragment 2-(Benzo[d]thiazol-2-yl)acetic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | (1,3-benzothiazol-2-yl)acetic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6CEC
 
 | | Crystal structure of fragment 3-(3-Methoxy-2-quinoxalinyl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(3-methoxyquinoxalin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Franzoni, I, Ravichandran, M, Lautens, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6CEF
 
 | | Crystal structure of fragment 3-(1,3-Benzothiazol-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(1,3-benzothiazol-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
7KIA
 
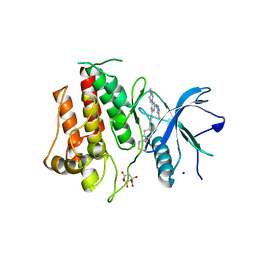 | | Crystal structure of FGFR2 kinase domain gatekeeper mutant V564F in complex with covalent compound 19 | | Descriptor: | 1-[4-(4-{4-(4-methylpiperazin-1-yl)-6-[(3-methyl-1H-pyrazol-5-yl)amino]pyrimidin-2-yl}phenyl)piperidin-1-yl]prop-2-en-1-one, CITRATE ANION, Fibroblast growth factor receptor 2, ... | | Authors: | Ke, J, Wibowo, A.S, Carter, J.J, Larsen, N.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of Aminopyrazole Derivatives as Potent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR2 and 3.
Acs Med.Chem.Lett., 12, 2021
|
|
7KIE
 
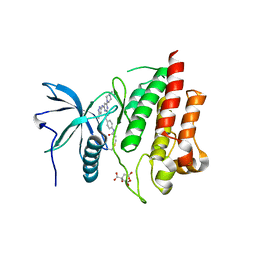 | | Crystal structure of FGFR2 kinase domain gatekeeper mutant V564F in complex with covalent compound 3 | | Descriptor: | CITRATE ANION, Fibroblast growth factor receptor 2, GLYCEROL, ... | | Authors: | Ke, J, Wibowo, A.S, Carter, J.J, Larsen, N.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery of Aminopyrazole Derivatives as Potent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR2 and 3.
Acs Med.Chem.Lett., 12, 2021
|
|
5KBQ
 
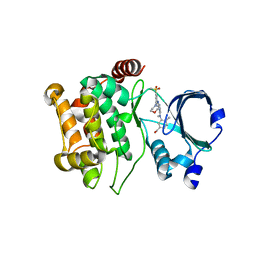 | | Pak1 in complex with bis-anilino pyrimidine inhibitor | | Descriptor: | Serine/threonine-protein kinase PAK 1, [4-methyl-3-[methyl-[2-[(3-methylsulfonyl-5-morpholin-4-yl-phenyl)amino]pyrimidin-4-yl]amino]phenyl]methanol | | Authors: | Ferguson, A. | | Deposit date: | 2016-06-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Optimization of Highly Kinase Selective Bis-anilino Pyrimidine PAK1 Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
6CN5
 
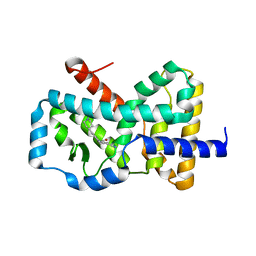 | |
6CN6
 
 | | RORC2 LBD complexed with compound 34 | | Descriptor: | 3-cyano-N-{3-[1-(cyclopentanecarbonyl)piperidin-4-yl]-1,4-dimethyl-1H-indol-5-yl}benzamide, Nuclear receptor ROR-gamma | | Authors: | Kauppi, B, Vajdos, F. | | Deposit date: | 2018-03-07 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of 3-Cyano- N-(3-(1-isobutyrylpiperidin-4-yl)-1-methyl-4-(trifluoromethyl)-1 H-pyrrolo[2,3- b]pyridin-5-yl)benzamide: A Potent, Selective, and Orally Bioavailable Retinoic Acid Receptor-Related Orphan Receptor C2 Inverse Agonist.
J. Med. Chem., 61, 2018
|
|
6SZE
 
 | |
8FDP
 
 | |
8F94
 
 | |
8FDR
 
 | | Structure of a dodecamer complex: 5'-CGCGAAAAGCCG-3-DB1476 | | Descriptor: | 4,4'-(1H-benzimidazole-2,6-diyl)di(benzene-1-carboximidamide), DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*TP*TP*TP*CP*GP*CP*G)-3'), ... | | Authors: | Ogbonna, E, Wilson, W.D. | | Deposit date: | 2022-12-04 | | Release date: | 2023-02-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
8FB4
 
 | | Structure of an alternating AT 16-mer bound by diamidine DB1476: 5'-GCTGGATATATCCAGC-3 | | Descriptor: | 4,4'-(1H-benzimidazole-2,6-diyl)di(benzene-1-carboximidamide), CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*GP*AP*TP*AP*TP*AP*TP*CP*CP*AP*GP*C)-3') | | Authors: | Terrell, J.R, Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-11-29 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
