7XFH
 
 | | Structure of nucleosome-AAG complex (A-30I, post-catalytic state) | | Descriptor: | DNA (152-MER), DNA-3-methyladenine glycosylase, Histone H2A type 1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
6TTP
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 1/Adenosine (DHU_M3M_023) | | Descriptor: | ACETATE ION, ADENOSINE, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
6K33
 
 | | Structure of PSI-isiA supercomplex from Thermosynechococcus vulcanus | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Akita, F, Nagao, R, Kato, K, Shen, J.R, Miyazaki, N. | | Deposit date: | 2019-05-16 | | Release date: | 2020-05-20 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structure of a cyanobacterial photosystem I surrounded by octadecameric IsiA antenna proteins.
Commun Biol, 3, 2020
|
|
7XFL
 
 | | Structure of nucleosome-AAG complex (A-53I, free state) | | Descriptor: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XNP
 
 | | Structure of nucleosome-AAG complex (A-55I, post-catalytic state) | | Descriptor: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFJ
 
 | | Structure of nucleosome-AAG complex (T-50I, post-catalytic state) | | Descriptor: | DNA (152-MER), DNA-3-methyladenine glycosylase, Histone H2A type 1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
1L05
 
 | |
6I3F
 
 | | Crystal structure of the complex of human angiotensinogen and renin at 2.55 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensinogen, ... | | Authors: | Yan, Y, Read, R.J. | | Deposit date: | 2018-11-06 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for the specificity of renin-mediated angiotensinogen cleavage.
J. Biol. Chem., 294, 2019
|
|
1L11
 
 | | CONTRIBUTIONS OF HYDROGEN BONDS OF THR 157 TO THE THERMODYNAMIC STABILITY OF PHAGE T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Dao-Pin, S, Wilson, K, Alber, T, Matthews, B.W. | | Deposit date: | 1988-02-05 | | Release date: | 1988-04-16 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme.
Nature, 330, 1987
|
|
1L14
 
 | |
1L46
 
 | |
1L65
 
 | |
6A6M
 
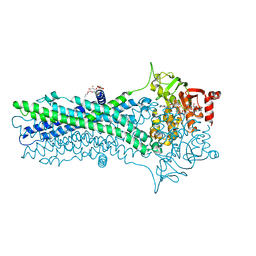 | | Crystal structure of an outward-open nucleotide-bound state of the eukaryotic ABC multidrug transporter CmABCB1 | | Descriptor: | ATP-binding cassette, sub-family B, member 1, ... | | Authors: | Kato, H, Nakatsu, T, Kodan, A. | | Deposit date: | 2018-06-28 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inward- and outward-facing X-ray crystal structures of homodimeric P-glycoprotein CmABCB1.
Nat Commun, 10, 2019
|
|
6KAF
 
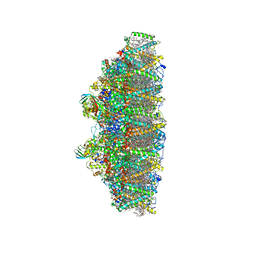 | | C2S2M2N2-type PSII-LHCII | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Chang, S.H, Shen, L.L, Huang, Z.H, Wang, W.D, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2019-06-22 | | Release date: | 2019-10-23 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure of a C2S2M2N2-type PSII-LHCII supercomplex from the green algaChlamydomonas reinhardtii.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1ULA
 
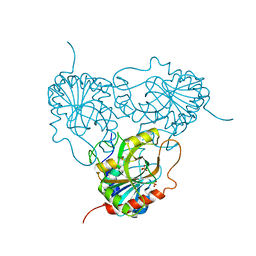 | | APPLICATION OF CRYSTALLOGRAPHIC AND MODELING METHODS IN THE DESIGN OF PURINE NUCLEOSIDE PHOSPHORYLASE INHIBITORS | | Descriptor: | PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Ealick, S.E, Rule, S.A, Carter, D.C, Greenhough, T.J, Babu, Y.S, Cook, W.J, Habash, J, Helliwell, J.R, Stoeckler, J.D, Parksjunior, R.E, Chen, S.-F, Bugg, C.E. | | Deposit date: | 1991-11-05 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of crystallographic and modeling methods in the design of purine nucleoside phosphorylase inhibitors.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1ULB
 
 | | APPLICATION OF CRYSTALLOGRAPHIC AND MODELING METHODS IN THE DESIGN OF PURINE NUCLEOSIDE PHOSPHORYLASE INHIBITORS | | Descriptor: | GUANINE, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Ealick, S.E, Rule, S.A, Carter, D.C, Greenhough, T.J, Babu, Y.S, Cook, W.J, Habash, J, Helliwell, J.R, Stoeckler, J.D, Parksjunior, R.E, Chen, S.-F, Bugg, C.E. | | Deposit date: | 1991-11-05 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of crystallographic and modeling methods in the design of purine nucleoside phosphorylase inhibitors.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1O6Y
 
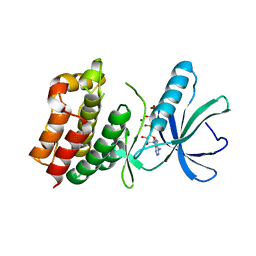 | | Catalytic domain of PknB kinase from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, SERINE/THREONINE-PROTEIN KINASE PKNB | | Authors: | Ortiz-Lombardia, M, Pompeo, F, Boitel, B, Alzari, P.M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-10-21 | | Release date: | 2003-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Pknb Serine/Threonine Kinase from Mycobacterium Tuberculosis
J.Biol.Chem., 278, 2003
|
|
1TMT
 
 | |
1A01
 
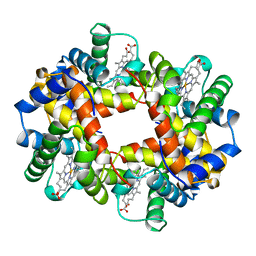 | | HEMOGLOBIN (VAL BETA1 MET, TRP BETA37 ALA) MUTANT | | Descriptor: | HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Arnone, A. | | Deposit date: | 1997-12-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution crystal structures of human hemoglobin with mutations at tryptophan 37beta: structural basis for a high-affinity T-state,.
Biochemistry, 37, 1998
|
|
5C8J
 
 | | A YidC-like protein in the archaeal plasma membrane | | Descriptor: | Antibody fragment, heavy chain, light chain, ... | | Authors: | Borowska, M.T, Dominik, P.K, Anghel, S.A, Kossiakoff, A.A, Keenan, R.J. | | Deposit date: | 2015-06-25 | | Release date: | 2015-09-02 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | A YidC-like Protein in the Archaeal Plasma Membrane.
Structure, 23, 2015
|
|
1L42
 
 | |
2H62
 
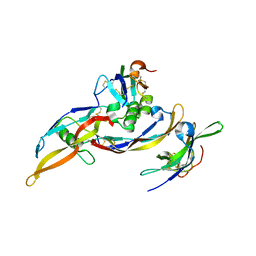 | | Crystal structure of a ternary ligand-receptor complex of BMP-2 | | Descriptor: | Acvr2b protein, Bone morphogenetic protein 2, Bone morphogenetic protein receptor type IA | | Authors: | Mueller, T.D. | | Deposit date: | 2006-05-30 | | Release date: | 2007-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A silent H-bond can be mutationally activated for high-affinity interaction of BMP-2 and activin type IIB receptor.
Bmc Struct.Biol., 7, 2007
|
|
3KQK
 
 | |
3B32
 
 | |
1SEP
 
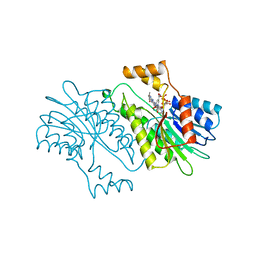 | | MOUSE SEPIAPTERIN REDUCTASE COMPLEXED WITH NADP AND SEPIAPTERIN | | Descriptor: | BIOPTERIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SEPIAPTERIN REDUCTASE | | Authors: | Auerbach, G, Herrmann, A, Guetlich, M, Fischer, M, Jacob, U, Bacher, A, Huber, R. | | Deposit date: | 1997-05-23 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The 1.25 A crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters.
EMBO J., 16, 1997
|
|
