1NEU
 
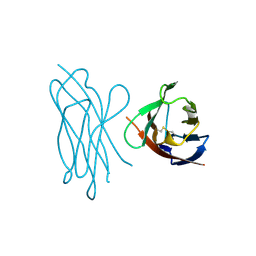 | | STRUCTURE OF MYELIN MEMBRANE ADHESION MOLECULE P0 | | Descriptor: | MYELIN P0 PROTEIN | | Authors: | Shapiro, L, Doyle, J.P, Hensley, P, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1996-09-24 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin.
Neuron, 17, 1996
|
|
1NEV
 
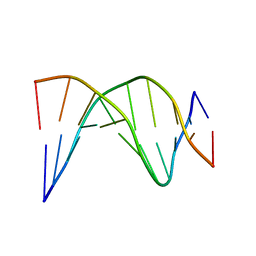 | | A-tract decamer | | Descriptor: | 5'-D(*CP*CP*GP*TP*TP*TP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*AP*AP*AP*CP*GP*G)-3' | | Authors: | Barbic, A, Zimmer, D.P, Crothers, D.M. | | Deposit date: | 2002-12-11 | | Release date: | 2003-03-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural origins of adenine-tract bending
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NEW
 
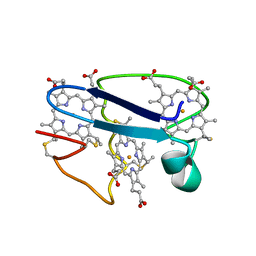 | | Cytochrome C551.5, NMR | | Descriptor: | CYTOCHROME C551.5, HEME C | | Authors: | Assfalg, M, Banci, L, Bertini, I, Bruschi, M, Turano, P. | | Deposit date: | 1998-02-10 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | 800 MHz 1H NMR solution structure refinement of oxidized cytochrome c7 from Desulfuromonas acetoxidans.
Eur.J.Biochem., 256, 1998
|
|
1NEX
 
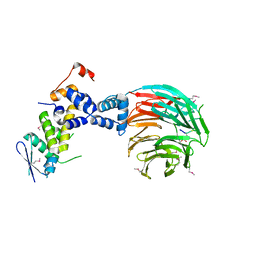 | | Crystal Structure of ScSkp1-ScCdc4-CPD peptide complex | | Descriptor: | CDC4 protein, Centromere DNA-binding protein complex CBF3 subunit D, GLL(TPO)PPQSG | | Authors: | Orlicky, S, Tang, X, Willems, A, Tyers, M, Sicheri, F. | | Deposit date: | 2002-12-12 | | Release date: | 2003-02-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Phosphodependent Substrate Selection and
Orientation by the SCFCdc4 Ubiquitin Ligase
Cell(Cambridge,Mass.), 112, 2003
|
|
1NEY
 
 | | Triosephosphate Isomerase in Complex with DHAP | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, triosephosphate isomerase | | Authors: | Jogl, G, Rozovsky, S, McDermott, A.E, Tong, L. | | Deposit date: | 2002-12-12 | | Release date: | 2003-01-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Optimal alignment for enzymatic proton transfer: Structure of the
Michaelis complex of triosephosphate isomerase at 1.2-A resolution.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NEZ
 
 | | The Crystal Structure of a TL/CD8aa Complex at 2.1A resolution:Implications for Memory T cell Generation, Co-receptor Preference and Affinity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, Y, Xiong, Y, Naidenko, O.V, Liu, J.H, Zhang, R, Joachimiak, A, Kronenberg, M, Cheroutre, H, Reinherz, E.L, Wang, J.H. | | Deposit date: | 2002-12-12 | | Release date: | 2003-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of a TL/CD8alphaalpha Complex at 2.1 A resolution: Implications for modulation of T cell activation and memory
Immunity, 18, 2003
|
|
1NF0
 
 | | Triosephosphate Isomerase in Complex with DHAP | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, triosephosphate isomerase | | Authors: | Jogl, G, Rozovsky, S, McDermott, A.E, Tong, L. | | Deposit date: | 2002-12-12 | | Release date: | 2003-01-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Optimal alignment for enzymatic proton transfer: Structure of the
Michaelis complex of triosephosphate isomerase at 1.2-A resolution
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NF1
 
 | | THE GAP RELATED DOMAIN OF NEUROFIBROMIN | | Descriptor: | PROTEIN (NEUROFIBROMIN) | | Authors: | Scheffzek, K, Ahmadian, M.R, Wiesmueller, L, Kabsch, W, Stege, P, Schmitz, F, Wittinghofer, A. | | Deposit date: | 1998-07-08 | | Release date: | 1999-07-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the GAP-related domain from neurofibromin and its implications.
EMBO J., 17, 1998
|
|
1NF2
 
 | |
1NF3
 
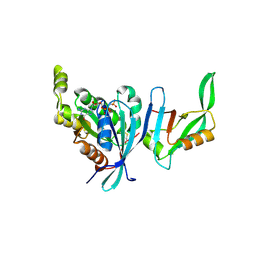 | | Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6 | | Descriptor: | G25K GTP-binding protein, placental isoform, MAGNESIUM ION, ... | | Authors: | Garrard, S.M, Capaldo, C.T, Gao, L, Rosen, M.K, Macara, I.G, Tomchick, D.R. | | Deposit date: | 2002-12-12 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6
Embo J., 22, 2003
|
|
1NF4
 
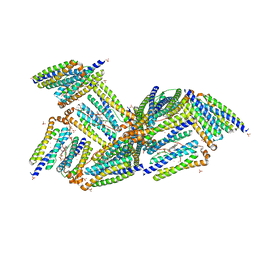 | | X-Ray Structure of the Desulfovibrio desulfuricans bacterioferritin: the diiron site in different states (reduced structure) | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, FE (II) ION, SULFATE ION, ... | | Authors: | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | Deposit date: | 2002-12-13 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
1NF5
 
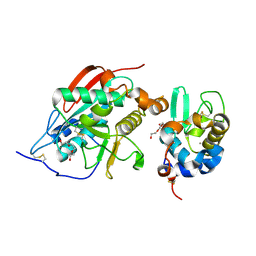 | | Crystal Structure of Lactose Synthase, Complex with Glucose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-lactalbumin, CALCIUM ION, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2002-12-13 | | Release date: | 2002-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Lactose Synthase Reveals a Large Conformational Change in its Catalytic Component, the beta-1,4-galactosyltransferase
J.Mol.Biol., 310, 2001
|
|
1NF6
 
 | | X-ray structure of the Desulfovibrio desulfuricans bacterioferritin: the diiron site in different catalytic states ("cycled" structure: reduced in solution and allowed to reoxidise before crystallisation) | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, FE (III) ION, GLYCEROL, ... | | Authors: | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | Deposit date: | 2002-12-13 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
1NF7
 
 | |
1NF8
 
 | | Crystal structure of PhzD protein active site mutant with substrate | | Descriptor: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, octyl beta-D-glucopyranoside, phenazine biosynthesis protein phzD | | Authors: | Parsons, F, Calabrese, K, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2002-12-13 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of Pseudomonas aeruginosa PhzD, an isochorismatase from the phenazine biosynthetic pathway
Biochemistry, 42, 2003
|
|
1NF9
 
 | | Crystal Structure of PhzD protein from Pseudomonas aeruginosa | | Descriptor: | FORMIC ACID, octyl beta-D-glucopyranoside, phenazine biosynthesis protein phzD | | Authors: | Parsons, F, Calabrese, K, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2002-12-13 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and mechanism of Pseudomonas aeruginosa PhzD, an isochorismatase from the phenazine biosynthetic pathway
Biochemistry, 42, 2003
|
|
1NFA
 
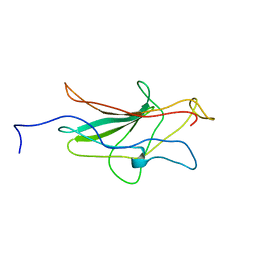 | | HUMAN TRANSCRIPTION FACTOR NFATC DNA BINDING DOMAIN, NMR, 10 STRUCTURES | | Descriptor: | HUMAN TRANSCRIPTION FACTOR NFATC1 | | Authors: | Wolfe, S.A, Zhou, P, Dotsch, V, Chen, L, You, A, Ho, S.N, Crabtree, G.R, Wagner, G, Verdine, G.L. | | Deposit date: | 1997-01-18 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Unusual Rel-like architecture in the DNA-binding domain of the transcription factor NFATc.
Nature, 385, 1997
|
|
1NFB
 
 | |
1NFD
 
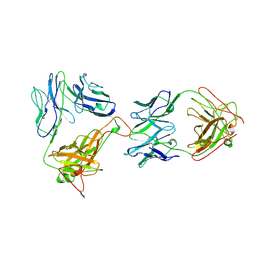 | | AN ALPHA-BETA T CELL RECEPTOR (TCR) HETERODIMER IN COMPLEX WITH AN ANTI-TCR FAB FRAGMENT DERIVED FROM A MITOGENIC ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H57 FAB, N15 ALPHA-BETA T-CELL RECEPTOR | | Authors: | Wang, J.-H, Lim, K, Smolyar, A, Teng, M.-K, Sacchittini, J, Reinherz, E.L. | | Deposit date: | 1997-08-04 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of an alphabeta T cell receptor (TCR) heterodimer in complex with an anti-TCR fab fragment derived from a mitogenic antibody.
EMBO J., 17, 1998
|
|
1NFF
 
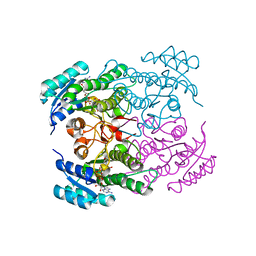 | | Crystal structure of Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-14 | | Release date: | 2002-12-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NFG
 
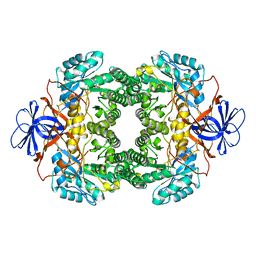 | | Structure of D-hydantoinase | | Descriptor: | D-hydantoinase, ZINC ION | | Authors: | Xu, Z, Yang, Y, Jiang, W, Arnold, E, Ding, J. | | Deposit date: | 2002-12-14 | | Release date: | 2003-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of D-Hydantoinase from Burkholderia pickettii at a Resolution of 2.7 Angstroms: Insights into the Molecular Basis of Enzyme Thermostability.
J.Bacteriol., 185, 2003
|
|
1NFH
 
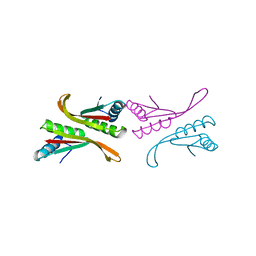 | | Structure of a Sir2 substrate, alba, reveals a mechanism for deactylation-induced enhancement of DNA-binding | | Descriptor: | conserved hypothetical protein AF1956 | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2002-12-15 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of a Sir2 substrate, Alba, reveals a mechanism for deacetylation-induced enhancement of DNA-binding
J.Biol.Chem., 278, 2003
|
|
1NFI
 
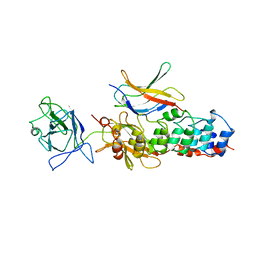 | | I-KAPPA-B-ALPHA/NF-KAPPA-B COMPLEX | | Descriptor: | I-KAPPA-B-ALPHA, NF-KAPPA-B P50, NF-KAPPA-B P65 | | Authors: | Jacobs, M.D, Harrison, S.C. | | Deposit date: | 1998-08-25 | | Release date: | 1998-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an IkappaBalpha/NF-kappaB complex.
Cell(Cambridge,Mass.), 95, 1998
|
|
1NFJ
 
 | | Structure of a Sir2 substrate, alba, reveals a mechanism for deactylation-induced enhancement of DNA-binding | | Descriptor: | conserved hypothetical protein AF1956 | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2002-12-15 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Sir2 substrate, alba, reveals a mechanism for deacetylation-induced enhancement of DNA-binding
J.Biol.Chem., 278, 2003
|
|
1NFK
 
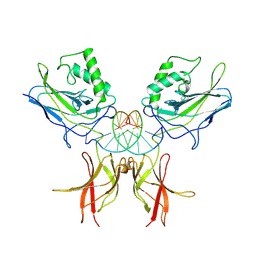 | | STRUCTURE OF THE NUCLEAR FACTOR KAPPA-B (NF-KB) P50 HOMODIMER | | Descriptor: | DNA (5'-D(*TP*GP*GP*GP*AP*AP*TP*TP*CP*CP*C)-3'), PROTEIN (NUCLEAR FACTOR KAPPA-B (NF-KB)) | | Authors: | Ghosh, G, Van Duyne, G, Ghosh, S, Sigler, P.B. | | Deposit date: | 1995-02-28 | | Release date: | 1996-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of NF-kappa B p50 homodimer bound to a kappa B site.
Nature, 373, 1995
|
|
