1TJW
 
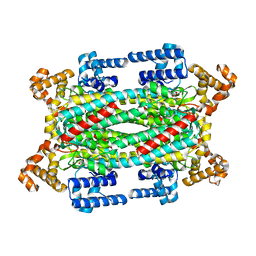 | | Crystal Structure of T161D Duck Delta 2 Crystallin Mutant with bound argininosuccinate | | Descriptor: | ARGININOSUCCINATE, Delta crystallin II | | Authors: | Sampaleanu, L.M, Codding, P.W, Lobsanov, Y.D, Tsai, M, Smith, G.D, Horvatin, C, Howell, P.L. | | Deposit date: | 2004-06-07 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of duck delta2 crystallin mutants provide insight into the role of Thr161 and the 280s loop in catalysis
Biochem.J., 384, 2004
|
|
1TXT
 
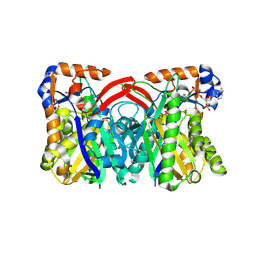 | | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase | | Descriptor: | 3-hydroxy-3-methylglutaryl-CoA synthase, ACETOACETYL-COENZYME A | | Authors: | Campobasso, N, Patel, M, Wilding, I.E, Kallender, H, Rosenberg, M, Gwynn, M. | | Deposit date: | 2004-07-06 | | Release date: | 2004-08-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism
J.Biol.Chem., 279, 2004
|
|
6MGR
 
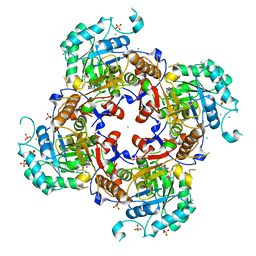 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor Oxanosine monophosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-[(Z)-(aminomethylidene)amino]-1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Yu, R, Hedstrom, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-14 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor Oxanosine Monophosphate
To Be Published
|
|
6JN8
 
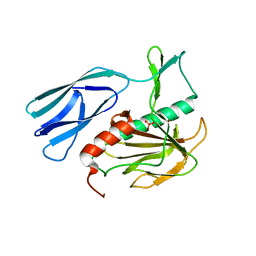 | | Structure of H216A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, SULFATE ION, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMX
 
 | | Structure of open form of peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, Peptidase M23, ... | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN1
 
 | | Structure of H247A mutant peptidoglycan peptidase complex with penta peptide | | Descriptor: | C0O-DAL-DAL, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMZ
 
 | | Structure of H247A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN0
 
 | | Structure of H247A mutant peptidoglycan peptidase complex with tetra-tri peptide | | Descriptor: | C0O-DAL-API, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.164 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN7
 
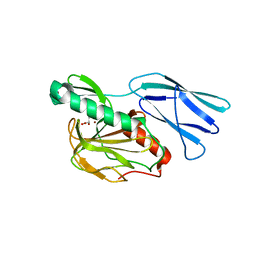 | | Structure of H216A mutant closed form peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMY
 
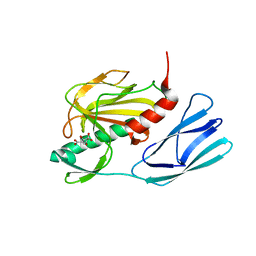 | | Structure of wild type closed form of peptidoglycan peptidase | | Descriptor: | CITRIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
3O7N
 
 | |
8T4C
 
 | |
5T1P
 
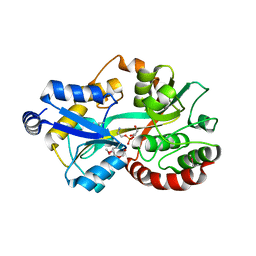 | | Crystal structure of the putative periplasmic solute-binding protein from Campylobacter jejuni | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC transporter, ... | | Authors: | Filippova, E.V, Wawrzsak, Z, Sandoval, J, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the putative periplasmic solute-binding protein from Campylobacter jejuni
To Be Published
|
|
6KV1
 
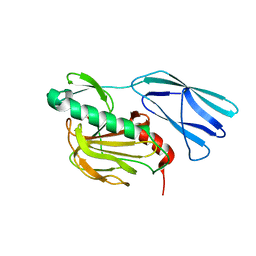 | | Structure of wild type closed form of peptidoglycan peptidase ZN SAD | | Descriptor: | CITRIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-09-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6KRT
 
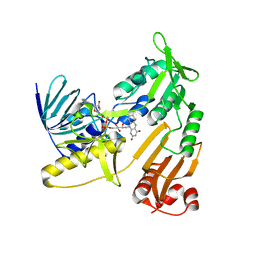 | | monodehydroascorbate reductase, MDHAR, from Antarctic hairgrass Deschampsia antarctica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, monodehydroascorbate reductase | | Authors: | Park, A.K, Do, H, Lee, J.H, Kim, H, Choi, W, Kim, I.S, Kim, H.W. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | monodehydroascorbate reductase, MDHAR, from Antarctic hairgrass Deschampsia antarctica
To Be Published
|
|
8SWD
 
 | |
6N4T
 
 | |
6HBV
 
 | |
6HBS
 
 | |
6HQI
 
 | | holo-form of polyphenol oxidase from Solanum lycopersicum | | Descriptor: | COPPER (II) ION, OXYGEN ATOM, Polyphenol oxidase A, ... | | Authors: | Kampatsikas, I, Bijelic, A, Rompel, A. | | Deposit date: | 2018-09-25 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural characterization of tomato polyphenol oxidases provide novel insights into their substrate specificity.
Sci Rep, 9, 2019
|
|
6HQJ
 
 | |
8TS0
 
 | |
6HMI
 
 | |
3NWF
 
 | | Glycoprotein B from Herpes simplex virus type 1, low-pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B, MESO-ERYTHRITOL, ... | | Authors: | Stampfer, S.D, Lou, H, Cohen, G.H, Eisenberg, R.J, Heldwein, E.E. | | Deposit date: | 2010-07-09 | | Release date: | 2010-12-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.
J.Virol., 84, 2010
|
|
3NYP
 
 | |
