3NY2
 
 | | Structure of the ubr-box of UBR2 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR2, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7KFA
 
 | | PCSK9 in complex with PCSK9i a 13mer cyclic peptide LDLR disruptor | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Chopra, R, Xu, M, Spraggon, G. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification of a PCSK9-LDLR disruptor peptide with in vivo function.
Cell Chem Biol, 29, 2022
|
|
2VTH
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H-pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design | | Descriptor: | 5-hydroxynaphthalene-1-sulfonamide, CELL DIVISION PROTEIN KINASE 2, GLYCEROL | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
6B6W
 
 | | Crystal structure of Desulfovibrio vulgaris carbon monoxide dehydrogenase, as-isolated (protein batch 2), oxidized C-cluster | | Descriptor: | CHLORIDE ION, Carbon monoxide dehydrogenase, FE(4)-NI(1)-S(4) CLUSTER, ... | | Authors: | Wittenborn, E.C, Drennan, C.L. | | Deposit date: | 2017-10-03 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Redox-dependent rearrangements of the NiFeS cluster of carbon monoxide dehydrogenase.
Elife, 7, 2018
|
|
8UZ5
 
 | |
7AEQ
 
 | | Human carbonic anhydrase II in complex with 2,3,5,6-tetrafluoro-4-(2-hydroxyethylsulfanyl)-N-methyl-benzenesulfonamide | | Descriptor: | 2,3,5,6-tetrakis(fluoranyl)-4-(2-hydroxyethylsulfanyl)-~{N}-methyl-benzenesulfonamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SODIUM ION, ... | | Authors: | Paketuryte, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and mechanism of secondary sulfonamide binding to carbonic anhydrases.
Eur.Biophys.J., 50, 2021
|
|
6PQZ
 
 | | P133G/S128A S. typhimurium siroheme synthase | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, Siroheme synthase | | Authors: | Pennington, J.M, Stroupe, M.E. | | Deposit date: | 2019-07-10 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Siroheme synthase orients substrates for dehydrogenase and chelatase activities in a common active site.
Nat Commun, 11, 2020
|
|
7KEZ
 
 | |
7AES
 
 | | Human carbonic anhydrase II in complex with 2,3,5,6-tetrafluoro-N-methyl-4-propylsulfanyl-benzenesulfonamide | | Descriptor: | 2,3,5,6-tetrakis(fluoranyl)-~{N}-methyl-4-propylsulfanyl-benzenesulfonamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BICINE, ... | | Authors: | Paketuryte, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and mechanism of secondary sulfonamide binding to carbonic anhydrases.
Eur.Biophys.J., 50, 2021
|
|
7ASZ
 
 | | L-2-haloacid dehalogenase H190A mutant from Zobellia galactanivorans | | Descriptor: | (S)-2-haloacid dehalogenase, PHOSPHATE ION, THIOCYANATE ION | | Authors: | Grigorian, E, Roret, T, Czjzek, M, Leblanc, C, Delage, L. | | Deposit date: | 2020-10-28 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | X-ray structure and mechanism of ZgHAD, a L-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans.
Protein Sci., 2022
|
|
7AGN
 
 | | Human carbonic anhydrase II in complex with 4-(2-aminoethylsulfanyl)-2,3,5,6-tetrafluoro-N-methyl-benzenesulfonamide | | Descriptor: | 4-(2-azanylethylsulfanyl)-2,3,5,6-tetrakis(fluoranyl)-~{N}-methyl-benzenesulfonamide, BICINE, SODIUM ION, ... | | Authors: | Paketuryte, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-09-23 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and mechanism of secondary sulfonamide binding to carbonic anhydrases.
Eur.Biophys.J., 50, 2021
|
|
8UUW
 
 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun12145 | | Descriptor: | 5-[2-(dimethylamino)ethoxy]-2-methyl-N-{(1R)-1-[(3P,5M)-3-(1-methyl-1H-pyrazol-4-yl)-5-(1,3-thiazol-5-yl)phenyl]ethyl}benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
7ARP
 
 | | Native L-2-haloacid dehalogenase from Zobellia galactanivorans | | Descriptor: | (S)-2-haloacid dehalogenase, PHOSPHATE ION, THIOCYANATE ION | | Authors: | Grigorian, E, Roret, T, Czjzek, M, Leblanc, C, Delage, L. | | Deposit date: | 2020-10-26 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-ray structure and mechanism of ZgHAD, a L-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans.
Protein Sci., 2022
|
|
7KF0
 
 | |
3J8I
 
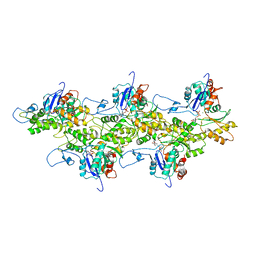 | | Near-Atomic Resolution for One State of F-Actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Galkin, V.E. | | Deposit date: | 2014-11-06 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Near-atomic resolution for one state of f-actin.
Structure, 23, 2015
|
|
6P5Z
 
 | |
5SV1
 
 | | Structure of the ExbB/ExbD complex from E. coli at pH 4.5 | | Descriptor: | Biopolymer transport protein ExbB, Biopolymer transport protein ExbD, MERCURY (II) ION | | Authors: | Celia, H, Botos, I, Lloubes, R, Buchanan, S.K, Noinaj, N. | | Deposit date: | 2016-08-04 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insight into the role of the Ton complex in energy transduction.
Nature, 538, 2016
|
|
3O6W
 
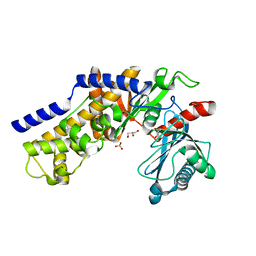 | | Crystal structure of monomeric KlHxk1 in crystal form VIII (open state) | | Descriptor: | GLYCEROL, Hexokinase, PHOSPHATE ION | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-29 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
7AGJ
 
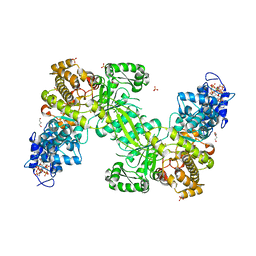 | | Ribonucleotide Reductase R1 protein from Aquifex aeolicus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Rehling, D, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2020-09-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Investigation of Class I Ribonucleotide Reductase from the Hyperthermophile Aquifex aeolicus.
Biochemistry, 61, 2022
|
|
2EIB
 
 | | Crystal Structure of Galactose Oxidase, W290H mutant | | Descriptor: | ACETATE ION, COPPER (II) ION, Galactose oxidase, ... | | Authors: | Phillips, S.E, McPherson, M.J, Knowles, P.F, Wilmot, C. | | Deposit date: | 2007-03-12 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Stacking Tryptophan of Galactose Oxidase: A Second-Coordination Sphere Residue that Has Profound Effects on Tyrosyl Radical Behavior and Enzyme Catalysis
Biochemistry, 46, 2007
|
|
2WAA
 
 | | Structure of a family two carbohydrate esterase from Cellvibrio japonicus | | Descriptor: | ACETATE ION, GLYCEROL, XYLAN ESTERASE, ... | | Authors: | Montainer, C, Money, V.A, Pires, V.M.R, Flint, J.E, Pinheiro, B.A, Goyal, A, Prates, J.A.M, Izumi, A, Stalbrand, H, Kolenova, K, Topakas, E, Dodson, E.J, Bolam, D.N, Davies, G.J, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-03-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
5IY0
 
 | | PfMCM N-terminal domain double hexamer | | Descriptor: | Cell division control protein 21, ZINC ION | | Authors: | Meagher, M, Enemark, E.J. | | Deposit date: | 2016-03-23 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a double hexamer of the Pyrococcus furiosus minichromosome maintenance protein N-terminal domain.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7KEV
 
 | | PCSK9 in complex with a cyclic peptide LDLR disruptor | | Descriptor: | CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, Proprotein convertase subtilisin/kexin type 9 Propeptide, ... | | Authors: | Spraggon, G, Chopra, R. | | Deposit date: | 2020-10-12 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a PCSK9-LDLR disruptor peptide with in vivo function.
Cell Chem Biol, 29, 2022
|
|
2J5D
 
 | | NMR structure of BNIP3 transmembrane domain in lipid bicelles | | Descriptor: | BCL2/ADENOVIRUS E1B 19 KDA PROTEIN-INTERACTING PROTEIN 3 | | Authors: | Bocharov, E.V, Pustovalova, Y.E, Volynsky, P.E, Maslennikov, I.V, Goncharuk, M.V, Ermolyuk, Y.S, Arseniev, A.S. | | Deposit date: | 2006-09-14 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger.
J. Biol. Chem., 282, 2007
|
|
5FMJ
 
 | | Bcl-xL with mouse Bak BH3 Q75L complex | | Descriptor: | 1,2-ETHANEDIOL, BAK1 PROTEIN, BCL-2-LIKE PROTEIN 1 | | Authors: | Fairlie, W.D, Lee, E.F, Smith, B.J. | | Deposit date: | 2015-11-06 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Physiological Restraint of Bak by Bcl-Xl is Essential for Cell Survival.
Genes Dev., 30, 2016
|
|
