4DIH
 
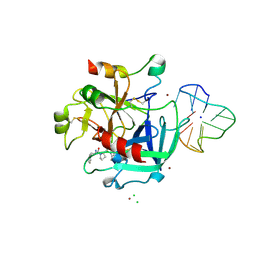 | | X-ray structure of the complex between human alpha thrombin and thrombin binding aptamer in the presence of sodium ions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2012-01-31 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity.
Nucleic Acids Res., 40, 2012
|
|
1V13
 
 | |
2ES2
 
 | |
1R49
 
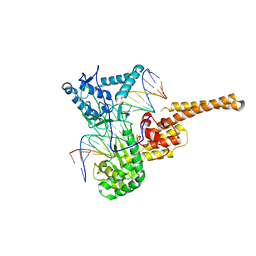 | | Human topoisomerase I (Topo70) double mutant K532R/Y723F | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*AP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(P*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*TP*AP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3', DNA topoisomerase I | | Authors: | Interthal, H, Quigley, P.M, Hol, W.G, Champoux, J.J. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | The role of lysine 532 in the catalytic mechanism of human topoisomerase I.
J.Biol.Chem., 279, 2004
|
|
3HJF
 
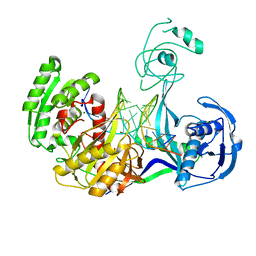 | | Crystal structure of T. thermophilus Argonaute E546 mutant protein complexed with DNA guide strand and 15-nt RNA target strand | | Descriptor: | 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', 5'-R(*CP*AP*AP*CP*CP*UP*AP*CP*UP*AP*CP*CP*UP*CP*G)-3', Argonaute, ... | | Authors: | Wang, Y, Li, H, Sheng, G, Patel, D.J. | | Deposit date: | 2009-05-21 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.056 Å) | | Cite: | Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes.
Nature, 461, 2009
|
|
6SH8
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 2, in the presence of ssDNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-08-06 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6SHB
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 1, in the presence of ssDNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-08-06 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
8S0A
 
 | | H. sapiens MCM2-7 hexamer bound to double stranded DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (22-mer), ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | MCM Double Hexamer Loading Visualised with Human Proteins
Nature, 2024
|
|
8S0D
 
 | | H. sapiens MCM bound to double stranded DNA and ORC1-6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (58-mer), DNA replication licensing factor MCM2, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | MCM Double Hexamer Loading Visualised with Human Proteins
Nature, 2024
|
|
8S09
 
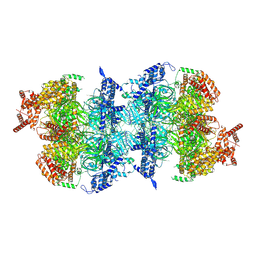 | | H. sapiens MCM2-7 double hexamer bound to double stranded DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (45-mer), ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | MCM Double Hexamer Loading Visualised with Human Proteins
Nature, 2024
|
|
8OX0
 
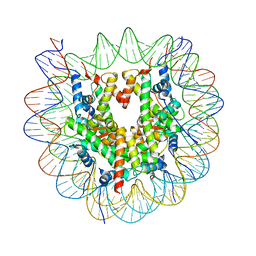 | | Structure of apo telomeric nucleosome | | Descriptor: | Histone H2A type 1-C, Histone H2B type 1-C/E/F/G/I, Histone H3.1, ... | | Authors: | Hu, H, van Roon, A.M.M, Ghanim, G.E, Ahsan, B, Oluwole, A, Peak-Chew, S, Robinson, C.V, Nguyen, T.H.D. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural basis of telomeric nucleosome recognition by shelterin factor TRF1.
Sci Adv, 9, 2023
|
|
2VPV
 
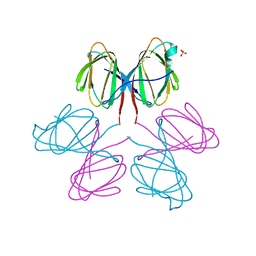 | | Dimerization Domain of Mif2p | | Descriptor: | PROTEIN MIF2, SULFATE ION | | Authors: | Cohen, R.L, Espelin, C.W, Sorger, P.K, Harrison, S.C, Simons, K.T. | | Deposit date: | 2008-03-05 | | Release date: | 2008-08-26 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Dissection of Mif2P, a Conserved DNA-Binding Kinetochore Protein.
Mol.Biol.Cell, 19, 2008
|
|
8CFA
 
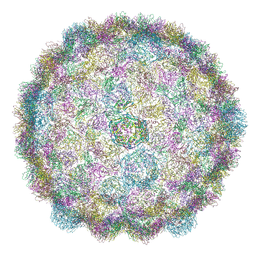 | |
6G1T
 
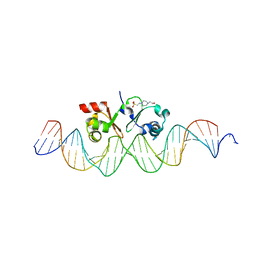 | | TraN, a repressor of an Enterococcus conjugative type IV secretion system | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AM32, DNA (34-MER) | | Authors: | Goessweiner-Mohr, N, Kohler, V, Keller, W. | | Deposit date: | 2018-03-22 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | TraN: A novel repressor of an Enterococcus conjugative type IV secretion system.
Nucleic Acids Res., 46, 2018
|
|
1RRJ
 
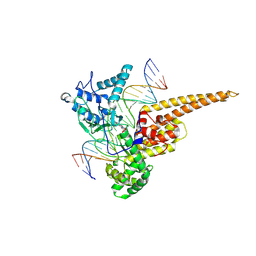 | | Structural Mechanisms of Camptothecin Resistance by Mutations in Human Topoisomerase I | | Descriptor: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, 2-(1-DIMETHYLAMINOMETHYL-2-HYDROXY-8-HYDROXYMETHYL-9-OXO-9,11-DIHYDRO-INDOLIZINO[1,2-B]QUINOLIN-7-YL)-2-HYDROXY-BUTYRIC ACID, 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T*GP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', ... | | Authors: | Chrencik, J.E, Staker, B.L, Burgin, A.B, Stewart, L, Redinbo, M.R. | | Deposit date: | 2003-12-08 | | Release date: | 2004-07-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of camptothecin resistance by human topoisomerase I mutations
J.Mol.Biol., 339, 2004
|
|
8CDY
 
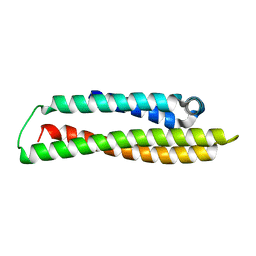 | | N-terminal domain of human apolipoprotein E | | Descriptor: | Maltodextrin-binding protein,Apolipoprotein E | | Authors: | Marek, M, Nemergut, M. | | Deposit date: | 2023-02-01 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Domino-like effect of C112R mutation on ApoE4 aggregation and its reduction by Alzheimer's Disease drug candidate.
Mol Neurodegener, 18, 2023
|
|
5A21
 
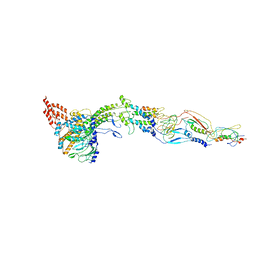 | | Structure of bacteriophage SPP1 head-to-tail interface without DNA and tape measure protein | | Descriptor: | 15 PROTEIN, HEAD COMPLETION PROTEIN GP16, MAJOR TAIL PROTEIN 17.1, ... | | Authors: | Chaban, Y, Lurz, R, Brasiles, S, Cornilleau, C, Karreman, M, Zinn-Justin, S, Tavares, P, Orlova, E.V. | | Deposit date: | 2015-05-06 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural Rearrangements in the Phage Head-to-Tail Interface During Assembly and Infection.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6K1I
 
 | | Human nucleosome core particle with gammaH2A.X variant | | Descriptor: | CHLORIDE ION, DNA (147-MER), Histone H2AX, ... | | Authors: | Sharma, D, De Falco, L, Davey, C.A. | | Deposit date: | 2019-05-10 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | PARP1 exhibits enhanced association and catalytic efficiency with gamma H2A.X-nucleosome.
Nat Commun, 10, 2019
|
|
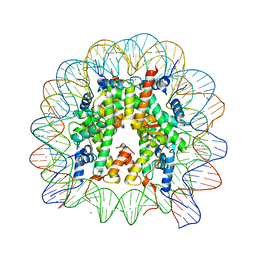
6K1K
 
 | | Human nucleosome core particle with H2A.X S139E variant | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2AX, ... | | Authors: | Sharma, D, De Falco, L, Davey, C.A. | | Deposit date: | 2019-05-10 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PARP1 exhibits enhanced association and catalytic efficiency with gamma H2A.X-nucleosome.
Nat Commun, 10, 2019
|
|
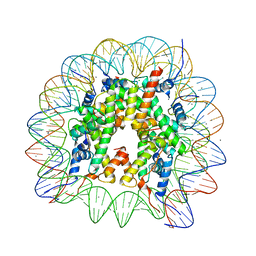
6K1J
 
 | | Human nucleosome core particle with H2A.X variant | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2AX, ... | | Authors: | Sharma, D, De Falco, L, Davey, C.A. | | Deposit date: | 2019-05-10 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | PARP1 exhibits enhanced association and catalytic efficiency with gamma H2A.X-nucleosome.
Nat Commun, 10, 2019
|
|
3X1L
 
 | |
4HQU
 
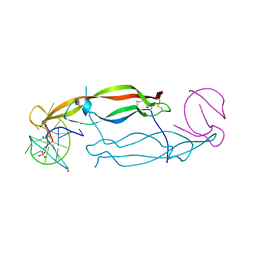 | | Crystal structure of human PDGF-BB in complex with a modified nucleotide aptamer (SOMAmer SL5) | | Descriptor: | MAGNESIUM ION, Platelet-derived growth factor subunit B, SODIUM ION, ... | | Authors: | Davies, D.R, Edwards, T.E, Janjic, N, Gelinas, A.D, Zhang, C, Jarvis, T.C. | | Deposit date: | 2012-10-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
352D
 
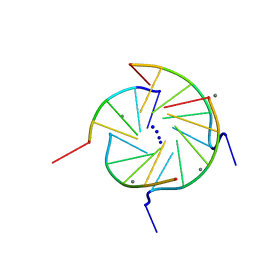 | | THE CRYSTAL STRUCTURE OF A PARALLEL-STRANDED PARALLEL-STRANDED GUANINE TETRAPLEX AT 0.95 ANGSTROM RESOLUTION | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3'), SODIUM ION | | Authors: | Phillips, K, Dauter, Z, Murchie, A.I.H, Lilley, D.M.J, Luisi, B. | | Deposit date: | 1997-09-04 | | Release date: | 1997-11-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The crystal structure of a parallel-stranded guanine tetraplex at 0.95 A resolution.
J.Mol.Biol., 273, 1997
|
|
2LXV
 
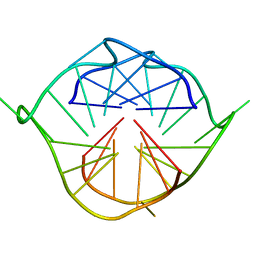 | |
7EEW
 
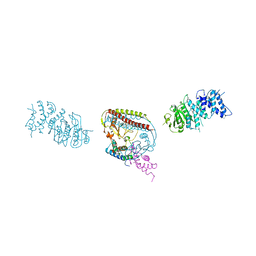 | |
