4C1P
 
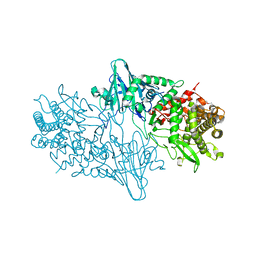 | | Geobacillus thermoglucosidasius GH family 52 xylosidase | | Descriptor: | BETA-XYLOSIDASE, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Espina, G, Eley, K, Schneider, T.R, Crennell, S.J, Danson, M.J. | | Deposit date: | 2013-08-13 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | A Novel Beta-Xylosidase Structure from Geobacillus Thermoglucosidasius: The First Crystal Structure of a Glycoside Hydrolase Family Gh52 Enzyme Reveals Unpredicted Similarity to Other Glycoside Hydrolase Folds
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CCX
 
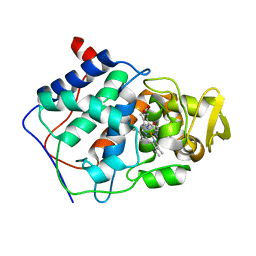 | | ALTERING SUBSTRATE SPECIFICITY AT THE HEME EDGE OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wilcox, S.K, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1995-03-17 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Altering substrate specificity at the heme edge of cytochrome c peroxidase.
Biochemistry, 35, 1996
|
|
7K2V
 
 | |
7K1W
 
 | |
6EYU
 
 | | Crystal structure of the inward H(+) pump xenorhodopsin | | Descriptor: | Bacteriorhodopsin, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Shevchenko, V, Polovinkin, V, Mager, T, Gushchin, I, Melnikov, I, Borshchevskiy, V, Popov, A, Alekseev, A, Gordeliy, V. | | Deposit date: | 2017-11-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inward H(+) pump xenorhodopsin: Mechanism and alternative optogenetic approach.
Sci Adv, 3, 2017
|
|
5IHL
 
 | |
2MW1
 
 | |
4E7D
 
 | |
2MQF
 
 | | NMR structure of spider toxin-TRTX-Hhn2b | | Descriptor: | Mu-theraphotoxin-Hhn2b | | Authors: | Klint, J.K, Chin, Y.K.Y, Mobli, M. | | Deposit date: | 2014-06-19 | | Release date: | 2015-07-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Rational Engineering Defines a Molecular Switch That Is Essential for Activity of Spider-Venom Peptides against the Analgesics Target NaV1.7
Mol.Pharmacol., 88, 2015
|
|
4EBK
 
 | | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, tobramycin-bound | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside nucleotidyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Dong, A, Minasov, G, Evdokimova, E, Egorova, O, Yim, V, Kudritska, M, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-23 | | Release date: | 2012-04-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, tobramycin-bound
To be Published
|
|
4EC0
 
 | | Crystal structure of hH-PGDS with water displacing inhibitor | | Descriptor: | 4-[2-(aminomethyl)naphthalen-1-yl]-N-[2-(morpholin-4-yl)ethyl]benzamide, GLUTATHIONE, Hematopoietic prostaglandin D synthase, ... | | Authors: | Day, J.E, Thorarensen, A, Trujillo, J.I. | | Deposit date: | 2012-03-26 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Investigation of the binding pocket of human hematopoietic prostaglandin (PG) D2 synthase (hH-PGDS): a tale of two waters.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4E7G
 
 | |
5IV0
 
 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with sulfonamide inhibitor 39 and coenzyme A | | Descriptor: | CHLORIDE ION, COENZYME A, Enhanced intracellular survival protein, ... | | Authors: | Gajadeera, C.S, Hou, C, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2016-03-18 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sulfonamide-Based Inhibitors of Aminoglycoside Acetyltransferase Eis Abolish Resistance to Kanamycin in Mycobacterium tuberculosis.
J. Med. Chem., 59, 2016
|
|
7K1Y
 
 | |
4ERL
 
 | |
1ZJ4
 
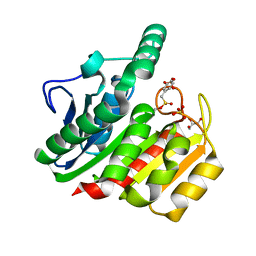 | | Crystal Structure Analysis of the dienelactone hydrolase mutant (E36D, C123S) bound with the PMS moiety of the protease inhibitor, Phenylmethylsulfonyl fluoride (PMSF)- 1.7 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-28 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5J58
 
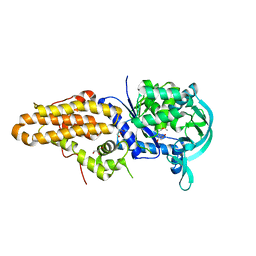 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor (Chem 1856) | | Descriptor: | (3S)-1-(5-chloro-1H-imidazo[4,5-b]pyridin-2-yl)-N-[(3,5-dichlorophenyl)methyl]piperidin-3-amine, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Barros-Alvarez, X, Hol, W.G.J. | | Deposit date: | 2016-04-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-guided design of novel Trypanosoma brucei Methionyl-tRNA synthetase inhibitors.
Eur J Med Chem, 124, 2016
|
|
4ED6
 
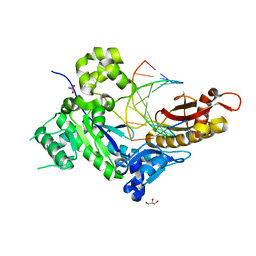 | | Human DNA polymerase eta - DNA ternary complex: Reaction in the AT crystal at pH 6.7 for 15 hr, Sideway translocation | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*TP*A)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
6FIY
 
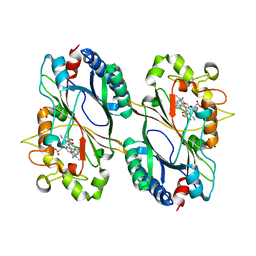 | | Crystal structure of a dye-decolorizing peroxidase D143AR232A variant from Klebsiella pneumoniae (KpDyP) | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Hofbauer, S, Mlynek, G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.09000432 Å) | | Cite: | Roles of distal aspartate and arginine of B-class dye-decolorizing peroxidase in heterolytic hydrogen peroxide cleavage.
J. Biol. Chem., 293, 2018
|
|
6EST
 
 | |
2NAO
 
 | | Atomic resolution structure of a disease-relevant Abeta(1-42) amyloid fibril | | Descriptor: | Beta-amyloid protein 42 | | Authors: | Waelti, M.A, Ravotti, F, Arai, H, Glabe, C, Wall, J, Bockmann, A, Guntert, P, Meier, B.H, Riek, R. | | Deposit date: | 2016-01-07 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Atomic-resolution structure of a disease-relevant A beta (1-42) amyloid fibril.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1W7B
 
 | |
5JFC
 
 | | NADH-dependent Ferredoxin:NADP Oxidoreductase (NfnI) from Pyrococcus furiosus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Zadvornyy, O.A, Schut, G.J, Nguyen, D.M, Artz, J.H, Tokmina-Lukaszewska, M, Lipscomb, G, King, P.W, Adams, M.W, Peters, J.W. | | Deposit date: | 2016-04-19 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Mechanistic insights into energy conservation by flavin-based electron bifurcation.
Nat. Chem. Biol., 13, 2017
|
|
7THF
 
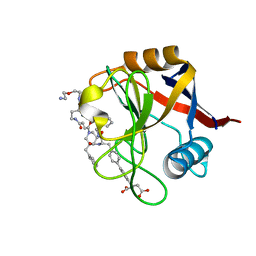 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B53 | | Descriptor: | 3-(4'-{[(4S,7S,11R,13E,19S)-19-{[2-(2-aminoethoxy)ethyl]carbamoyl}-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosin-4-yl]methyl}[1,1'-biphenyl]-4-yl)pentanedioic acid, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7TGV
 
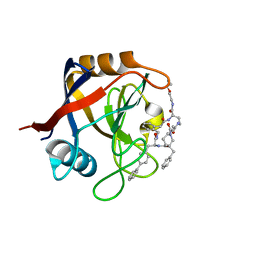 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B2 | | Descriptor: | (4S,7S,11R,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-7-benzyl-4-[([1,1'-biphenyl]-4-yl)methyl]-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
