5UGJ
 
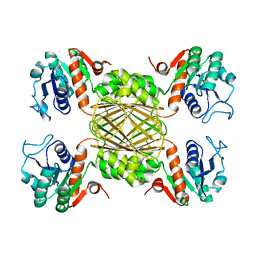 | |
7VRR
 
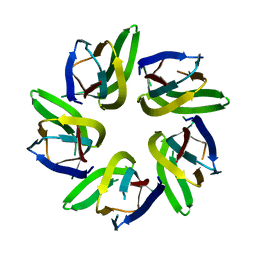 | |
7Q24
 
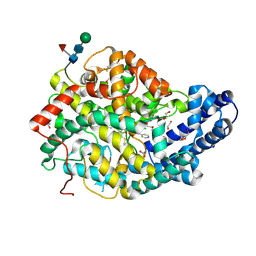 | |
5D3F
 
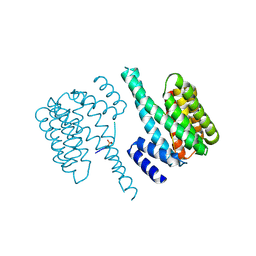 | |
7NA0
 
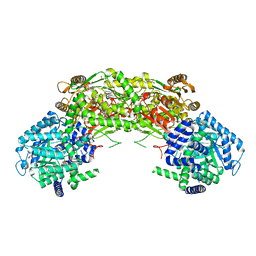 | |
7Q25
 
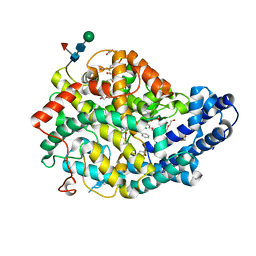 | |
8V9N
 
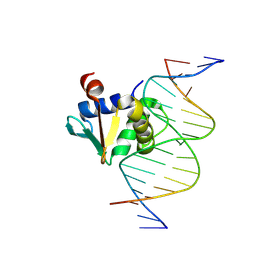 | |
5HK5
 
 | |
8ONX
 
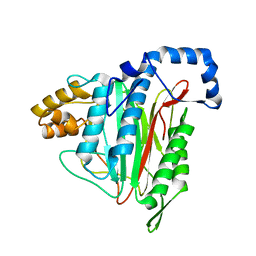 | | High resolution structure of Chaetomium thermophilum MAP2 | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
7Q26
 
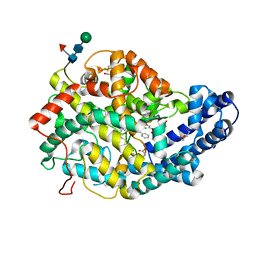 | | Crystal structure of Angiotensin-1 converting enzyme N-domain in complex with dual ACE/NEP inhibitor AD013 | | Descriptor: | (2~{S},5~{R})-5-(4-methylphenyl)-1-[2-[[(2~{S})-1-oxidanyl-1-oxidanylidene-4-phenyl-butan-2-yl]amino]ethanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2021-10-23 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the Requirements for Dual Angiotensin-Converting Enzyme C-Domain Selective/Neprilysin Inhibition.
J.Med.Chem., 65, 2022
|
|
8V35
 
 | | Crystal structure of HpsN from Cupriavidus pinatubonensis | | Descriptor: | 1,2-ETHANEDIOL, Sulfopropanediol 3-dehydrogenase, ZINC ION | | Authors: | Lee, M. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and kinetic insights into the stereospecific oxidation of R -2,3-dihydroxypropanesulfonate by DHPS-3-dehydrogenase from Cupriavidus pinatubonensis.
Chem Sci, 15, 2024
|
|
8RD5
 
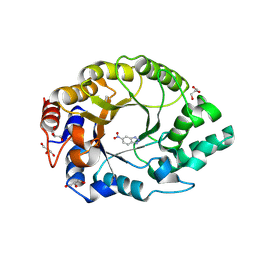 | | Crystal structure of Kemp Eliminase HG3.R5 with bound transition state analog 6-nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, ACETATE ION, Endo-1,4-beta-xylanase, ... | | Authors: | Schaub, D, Schwander, T, Hueppi, S, Buller, R.M. | | Deposit date: | 2023-12-07 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enriching productive mutational paths accelerates enzyme evolution.
Nat.Chem.Biol., 2024
|
|
7Q27
 
 | |
8U59
 
 | |
7ZC8
 
 | |
8U9H
 
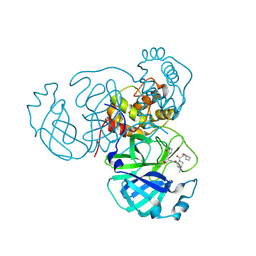 | |
8OGE
 
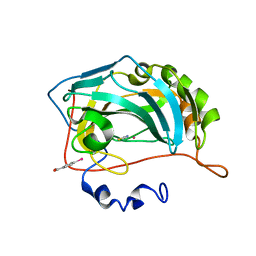 | | Structure of cobalt(II) substituted double mutant human carbonic anhydrase II bound to thiocyanate | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, COBALT (II) ION, Carbonic anhydrase 2, ... | | Authors: | Silva, J.M, Cerofolini, L, Carvalho, A.L, Ravera, E, Fragai, M, Parigi, G, Macedo, A.L, Geraldes, C.F.G.C, Luchinat, C. | | Deposit date: | 2023-03-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Elucidating the concentration-dependent effects of thiocyanate binding to carbonic anhydrase.
J.Inorg.Biochem., 244, 2023
|
|
8U9U
 
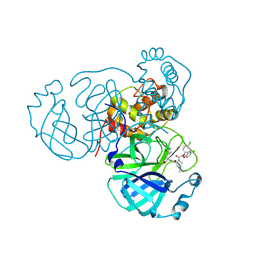 | |
7YZE
 
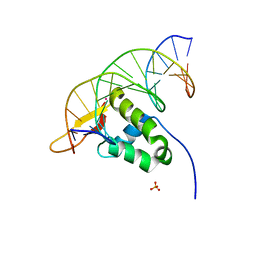 | |
5HKE
 
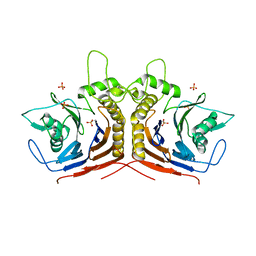 | | bile salt hydrolase from Lactobacillus salivarius | | Descriptor: | Bile salt hydrolase, PHOSPHATE ION | | Authors: | Hu, X.-J. | | Deposit date: | 2016-01-14 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of bile salt hydrolase from Lactobacillus salivarius.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
6MKI
 
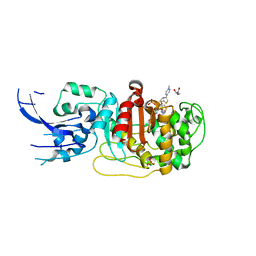 | | Crystal structure of penicillin-binding protein 4 (PBP4) from Enterococcus faecalis in the ceftaroline-bound form | | Descriptor: | Ceftaroline, bound form, GLYCEROL, ... | | Authors: | D'Andrea, E.D, Moon, T.M, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
5HKK
 
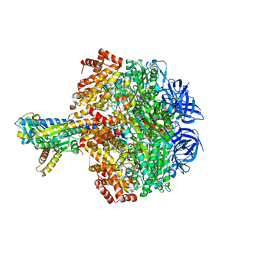 | | Caldalaklibacillus thermarum F1-ATPase (wild type) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Ferguson, S.A, Cook, G.M, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2016-01-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Regulation of the thermoalkaliphilic F1-ATPase from Caldalkalibacillus thermarum.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7VR6
 
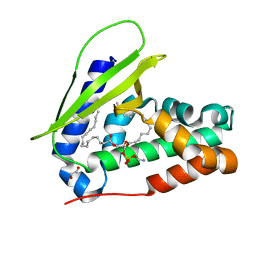 | | Crystal structure of MlaC from Escherichia coli in quasi-open state | | Descriptor: | 1,2-ETHANEDIOL, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Intermembrane phospholipid transport system binding protein MlaC | | Authors: | Dutta, A, Kanaujia, S.P. | | Deposit date: | 2021-10-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MlaC belongs to a unique class of non-canonical substrate-binding proteins and follows a novel phospholipid-binding mechanism.
J.Struct.Biol., 214, 2022
|
|
7Z5W
 
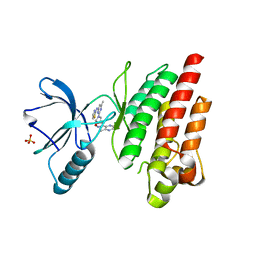 | | ROS1 with AstraZeneca ligand 1 | | Descriptor: | Proto-oncogene tyrosine-protein kinase ROS, SULFATE ION, ~{N}-[6-methyl-2-[(2~{S})-2-[3-(3-methylpyrazin-2-yl)-1,2-oxazol-5-yl]pyrrolidin-1-yl]pyrimidin-4-yl]-1,3-thiazol-2-amine | | Authors: | Hargreaves, D. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Virtual Screening in the Cloud Identifies Potent and Selective ROS1 Kinase Inhibitors.
J.Chem.Inf.Model., 62, 2022
|
|
6ML7
 
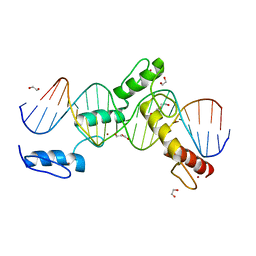 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpG 5mC Modification) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*(5CM)P*GP*AP*AP*TP*T)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
