4PMT
 
 | |
4PMP
 
 | |
4PEQ
 
 | | Structure of bovine ribonuclease inhibitor complexed with bovine ribonuclease I | | Descriptor: | Ribonuclease pancreatic, Ribonuclease/angiogenin inhibitor 1 | | Authors: | Bianchetti, C.M, Lomax, J.E, Raines, R.T, Fox, B.G. | | Deposit date: | 2014-04-24 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Functional evolution of ribonuclease inhibitor: insights from birds and reptiles.
J.Mol.Biol., 426, 2014
|
|
4PER
 
 | | Structure of Gallus gallus ribonuclease inhibitor complexed with Gallus gallus ribonuclease I | | Descriptor: | Angiogenin, Ribonuclease Inhibitor | | Authors: | Bianchetti, C.M, Lomax, J.E, Raines, R.T, Fox, B.G. | | Deposit date: | 2014-04-24 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Functional evolution of ribonuclease inhibitor: insights from birds and reptiles.
J.Mol.Biol., 426, 2014
|
|
3TSR
 
 | | X-ray structure of mouse ribonuclease inhibitor complexed with mouse ribonuclease 1 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ribonuclease inhibitor, ... | | Authors: | Chang, A, Lomax, J.E, Bingman, C.A, Raines, R.T, Phillips Jr, G.N. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1999 Å) | | Cite: | Functional evolution of ribonuclease inhibitor: insights from birds and reptiles.
J.Mol.Biol., 426, 2014
|
|
5T1Z
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with Ethoxytriphenylethylene and GRIP Peptide | | Descriptor: | 4,4'-[(1Z)-1-(4-ethoxyphenyl)but-1-ene-1,2-diyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Fanning, S.W, Rajan, S.S, Maximov, P.Y, Abderrahman, B.H, Surojeet, S, Fernandes, D.J, Fan, P, Curpan, R.F, Greene, G.L, Jordan, V.C. | | Deposit date: | 2016-08-22 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Endoxifen, 4-Hydroxytamoxifen and an Estrogenic Derivative Modulate Estrogen Receptor Complex Mediated Apoptosis in Breast Cancer.
Mol. Pharmacol., 94, 2018
|
|
3JZ7
 
 | |
2ON8
 
 | |
2ONQ
 
 | |
3FIL
 
 | |
5HYW
 
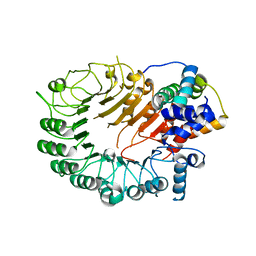 | | The crystal structure of the D3-ASK1 complex | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | Deposit date: | 2016-02-02 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
2LW5
 
 | |
2KX4
 
 | | Solution structure of Bacteriophage Lambda gpFII | | Descriptor: | Tail attachment protein | | Authors: | Maxwell, K.L, Cardarelli, L, Neudecker, P, Davidson, A.R, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2010-04-26 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Phages have adapted the same protein fold to fulfill multiple functions in virion assembly.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3UKX
 
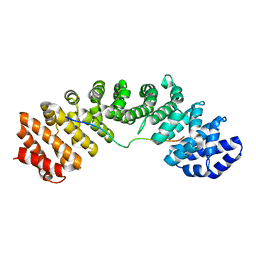 | | Mouse importin alpha: Bimax2 peptide complex | | Descriptor: | Bimax2 peptide, Importin subunit alpha-2 | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
3KAK
 
 | | Structure of homoglutathione synthetase from Glycine max in open conformation with gamma-glutamyl-cysteine bound. | | Descriptor: | GAMMA-GLUTAMYLCYSTEINE, Homoglutathione synthetase | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
3KAL
 
 | | Structure of homoglutathione synthetase from Glycine max in closed conformation with homoglutathione, ADP, a sulfate ion, and three magnesium ions bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-gamma-glutamyl-L-cysteinyl-beta-alanine, MAGNESIUM ION, ... | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
6Y4C
 
 | | Structure of galectin-3C in complex with lactose determined by serial crystallography using an XtalTool support | | Descriptor: | CHLORIDE ION, Galectin-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shilova, A, Hakansson, M, Welin, M, Kovacic, R, Mueller, U, Logan, D.T. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Current status and future opportunities for serial crystallography at MAX IV Laboratory.
J.Synchrotron Radiat., 27, 2020
|
|
6Y2N
 
 | | Crystal structure of ribonucleotide reductase R2 subunit solved by serial synchrotron crystallography | | Descriptor: | FE (III) ION, MANGANESE (III) ION, Ribonucleoside-diphosphate reductase subunit beta | | Authors: | Shilova, A, Lebrette, H, Aurelius, O, Hogbom, M, Mueller, U. | | Deposit date: | 2020-02-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Current status and future opportunities for serial crystallography at MAX IV Laboratory.
J.Synchrotron Radiat., 27, 2020
|
|
6Y78
 
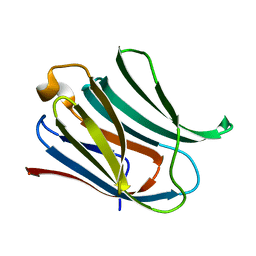 | | Structure of galectin-3C in complex with lactose determined by serial crystallography using a silicon nitride membrane support | | Descriptor: | Galectin-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hakansson, M, Welin, M, Shilova, A, Kovacic, R, Mueller, U, Logan, D.T. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Current status and future opportunities for serial crystallography at MAX IV Laboratory.
J.Synchrotron Radiat., 27, 2020
|
|
6R5J
 
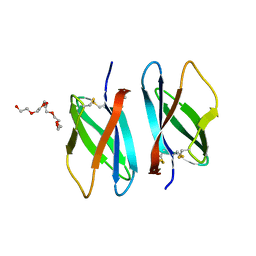 | |
5ZNE
 
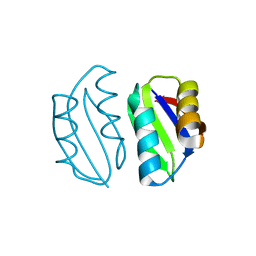 | |
5ZNG
 
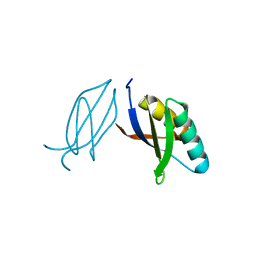 | | The crystal complex of immune receptor RGA5A_S of Pia from rice (Oryzae sativa) with rice blast (Magnaporthe oryzae) effector protein AVR1-CO39 | | Descriptor: | AVR1-CO39, NBS-LRR type protein | | Authors: | Guo, L.W, Zhang, Y.K, Liu, Q, Ma, M.Q, Liu, J.F, Peng, Y.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Specific recognition of two MAX effectors by integrated HMA domains in plant immune receptors involves distinct binding surfaces
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7W6K
 
 | | Cryo-EM structure of GmALMT12/QUAC1 anion channel | | Descriptor: | GmALMT12/QUAC1 | | Authors: | Qin, L, Tang, L.H, Xu, J.S, Zhang, X.H, Zhu, Y, Sun, F, Su, M, Zhai, Y.J, Chen, Y.H. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure and electrophysiological characterization of ALMT from Glycine max reveal a previously uncharacterized class of anion channels.
Sci Adv, 8, 2022
|
|
3KAJ
 
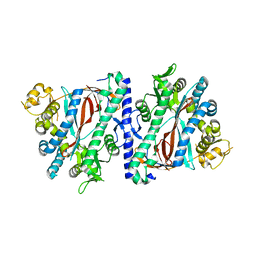 | | Apoenzyme structure of homoglutathione synthetase from Glycine max in open conformation | | Descriptor: | Homoglutathione synthetase | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
7CKA
 
 | |
