8FBF
 
 | | Crystal structure of OrfX2 from Clostridium botulinum E1 | | Descriptor: | Neurotoxin complex component Orf-X2, SULFATE ION | | Authors: | Lam, K.H, Gao, L. | | Deposit date: | 2022-11-29 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of OrfX1, OrfX2 and the OrfX1-OrfX3 complex from the orfX gene cluster of botulinum neurotoxin E1.
Febs Lett., 597, 2023
|
|
8FBD
 
 | | Crystal structure of OrfX1-OrfX3 complex from Clostridium botulinum E1 | | Descriptor: | ACETATE ION, Neurotoxin complex component Orf-X1, Neurotoxin complex component Orf-X3 | | Authors: | Liu, S, Gao, L. | | Deposit date: | 2022-11-29 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of OrfX1, OrfX2 and the OrfX1-OrfX3 complex from the orfX gene cluster of botulinum neurotoxin E1.
Febs Lett., 597, 2023
|
|
8FBE
 
 | | Crystal structure of OrfX1 from Clostridium botulinum E1 | | Descriptor: | Neurotoxin complex component Orf-X1 | | Authors: | Liu, S, Gao, L. | | Deposit date: | 2022-11-29 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structures of OrfX1, OrfX2 and the OrfX1-OrfX3 complex from the orfX gene cluster of botulinum neurotoxin E1.
Febs Lett., 597, 2023
|
|
3TV6
 
 | | Human B-Raf Kinase Domain in Complex with a Methoxypyrazolopyridinyl Benzamide Inhibitor | | Descriptor: | 2,6-difluoro-N-(3-methoxy-2H-pyrazolo[3,4-b]pyridin-5-yl)-3-[(propylsulfonyl)amino]benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Voegtli, W.C, Sturgis, H.L, Wu, W.-I. | | Deposit date: | 2011-09-19 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Pyrazolopyridine Inhibitors of B-Raf(V600E). Part 1: The Development of Selective, Orally Bioavailable, and Efficacious Inhibitors.
ACS Med Chem Lett, 2, 2011
|
|
1E0E
 
 | | N-terminal zinc-binding HHCC domain of HIV-2 integrase | | Descriptor: | HUMAN IMMUNODEFICIENCY VIRUS TYPE 2 INTEGRASE, ZINC ION | | Authors: | Eijkelenboom, A.P.A.M, Van Den ent, F.M.I, Plasterk, R.H.A, Kaptein, R, Boelens, R. | | Deposit date: | 2000-03-25 | | Release date: | 2001-03-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Refined Solution Structure of the Dimeric N-Terminal Hhcc Domain of HIV-2 Integrase
J.Biomol.NMR, 18, 2000
|
|
5G1W
 
 | | Apo Structure of Linalool Dehydratase-Isomerase | | Descriptor: | 1,2-ETHANEDIOL, LINALOOL DEHYDRATASE/ISOMERASE, METHYLMALONIC ACID | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
5G1U
 
 | | Linalool Dehydratase Isomerase in complex with Geraniol | | Descriptor: | Geraniol, LINALOOL DEHYDRATASE/ISOMERASE | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
5G1V
 
 | | Linalool Dehydratase Isomerase: Selenomethionine Derivative | | Descriptor: | LINALOOL DEHYDRATASE ISOMERASE | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
8H7A
 
 | | Crystal structure of the dimer form KAT6A WH domain with its bound double stranded DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*CP*GP*AP*CP*GP*GP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*CP*CP*GP*TP*CP*GP*GP*AP*CP*C)-3'), Histone acetyltransferase KAT6A, ... | | Authors: | Wang, Z, Cao, Y. | | Deposit date: | 2022-10-19 | | Release date: | 2023-01-18 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The histone acetyltransferase KAT6A is recruited to unmethylated CpG islands via a DNA binding winged helix domain.
Nucleic Acids Res., 51, 2023
|
|
9ARL
 
 | |
9ARJ
 
 | |
9ARK
 
 | |
9ATV
 
 | |
5IFW
 
 | |
2KB6
 
 | | Solution structure of onconase C87A/C104A | | Descriptor: | Protein P-30 | | Authors: | Weininger, U, Schulenburg, C, Arnold, U, Ulbrich-Hofmann, R, Balbach, J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Impact of the C-terminal disulfide bond on the folding and stability of onconase.
Chembiochem, 11, 2010
|
|
4EIG
 
 | | CA1698 camel antibody fragment in complex with DHFR | | Descriptor: | CA1698 camel antibody fragment, Dihydrofolate reductase | | Authors: | Oyen, D, Srinivasan, V. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihyrofolate reductase of Escherichia coli.
Biochim.Biophys.Acta, 1834, 2013
|
|
4EIZ
 
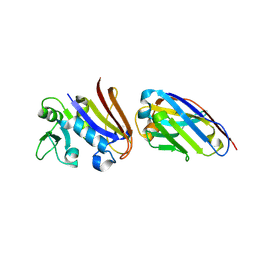 | | Structure of Nb113 bound to apoDHFR | | Descriptor: | Dihydrofolate reductase, Nb113 Camel antibody fragment | | Authors: | Oyen, D, Srinivasan, V. | | Deposit date: | 2012-04-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihyrofolate reductase of Escherichia coli.
Biochim.Biophys.Acta, 1834, 2013
|
|
2LFJ
 
 | |
4FHB
 
 | | Enhancing DHFR catalysis by binding of an allosteric regulator nanobody (Nb179) | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Oyen, D. | | Deposit date: | 2012-06-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihyrofolate reductase of Escherichia coli.
Biochim.Biophys.Acta, 1834, 2013
|
|
4FGV
 
 | |
4EJ1
 
 | |
1O58
 
 | |
1O2D
 
 | |
7BL5
 
 | | pre-50S-ObgE particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T, Schmidt, S. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL4
 
 | | in vitro reconstituted 50S-ObgE-GMPPNP-RsfS particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
