8TDQ
 
 | | SFX-XFEL structure of CYP121 cocrystallized with substrate cYY | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Nguyen, R.C, Dasgupta, M, Bhowmick, A, Kern, J.F, Liu, A. | | Deposit date: | 2023-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | In Situ Structural Observation of a Substrate- and Peroxide-Bound High-Spin Ferric-Hydroperoxo Intermediate in the P450 Enzyme CYP121.
J.Am.Chem.Soc., 145, 2023
|
|
8TDP
 
 | | Time-resolved SFX-XFEL crystal structure of CYP121 bound with cYY reacted with peracetic acid for 200 milliseconds | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, HYDROGEN PEROXIDE, Mycocyclosin synthase, ... | | Authors: | Nguyen, R.C, Dasgupta, M, Bhowmick, A, Kern, J.F, Liu, A. | | Deposit date: | 2023-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In Situ Structural Observation of a Substrate- and Peroxide-Bound High-Spin Ferric-Hydroperoxo Intermediate in the P450 Enzyme CYP121.
J.Am.Chem.Soc., 145, 2023
|
|
8FC5
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and azithromycin at 2.65A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
8FC4
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and erythromycin at 2.45A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
8FC6
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and telithromycin at 2.35A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
8FC3
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and telithromycin at 2.60A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
8FC2
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and azithromycin at 2.50A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
8TRD
 
 | | mGluR3 class 1 in the presence of the antagonist LY 341495 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, Metabotropic glutamate receptor 3 | | Authors: | Strauss, A, Levitz, J. | | Deposit date: | 2023-08-09 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of positive allosteric modulation of metabotropic glutamate receptor activation and internalization.
Nat Commun, 15, 2024
|
|
8TRC
 
 | |
8TR2
 
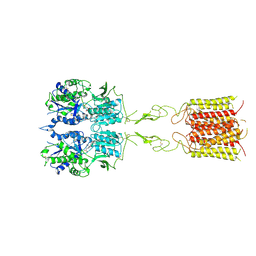 | | mGluR3 in the presence of the agonist LY379268 | | Descriptor: | (1S,4R,5R,6S)-4-amino-2-oxabicyclo[3.1.0]hexane-4,6-dicarboxylic acid, CALCIUM ION, Metabotropic glutamate receptor 3 | | Authors: | Strauss, A, Levitz, J. | | Deposit date: | 2023-08-09 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of allosteric modulation of metabotropic glutamate receptor activation and desensitization.
Biorxiv, 2023
|
|
8TQB
 
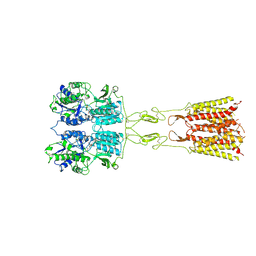 | | mGluR3 in the presence of the agonist LY379268 and PAM VU6023326 | | Descriptor: | (1R,4R,5S,6R)-4-azanyl-2-oxabicyclo[3.1.0]hexane-4,6-dicarboxylic acid, CALCIUM ION, Metabotropic glutamate receptor 3 | | Authors: | Strauss, A, Levitz, J. | | Deposit date: | 2023-08-06 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of allosteric modulation of metabotropic glutamate receptor activation and desensitization.
Biorxiv, 2023
|
|
8TR0
 
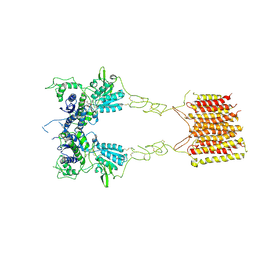 | |
8FC1
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and erythromycin at 2.50A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
8UEE
 
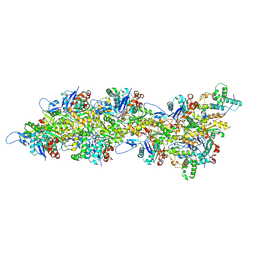 | | Atomic structure of Salmonella SipA/F-actin complex by cryo-EM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Niedzialkowska, E, Runyan, L, Kudryashova, E, Kudryashov, D.S, Egelman, E.H. | | Deposit date: | 2023-10-01 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Stabilization of F-actin by Salmonella effector SipA resembles the structural effects of inorganic phosphate and phalloidin.
Structure, 32, 2024
|
|
8V5W
 
 | | UIC-1 mutant UIC-1-B5T | | Descriptor: | UIC-1-B5T | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
8V61
 
 | | UIC-1 mutant - UIC-1-L6I | | Descriptor: | UIC-1-L6I | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
8V5X
 
 | | UIC-1 mutant - UIC-1-L6A | | Descriptor: | ACETONITRILE, UIC-1-L6A | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
8V59
 
 | | UIC-1 mutant - UIC-1-B5I | | Descriptor: | METHANOL, UIC-1-B5I | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-11-30 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
8V5Z
 
 | | UIC-1 mutant - UIC-1-L6M | | Descriptor: | ACETONITRILE, UIC-1-L6M | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
8V56
 
 | | UIC-1 mutant - UIC-1-B5W | | Descriptor: | UIC-1-B5W | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-11-30 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
8V4N
 
 | | Myxococcus xanthus EncA 3xHis pore mutant with T=1 icosahedral symmetry | | Descriptor: | Type 1 encapsulin shell protein EncA | | Authors: | Szyszka, T.N, Andreas, M.P, Lie, F, Miller, L.M, Adamson, L.S.R, Fatehi, F, Twarock, R, Draper, B.E, Jarrold, M.F, Giessen, T.W, Lau, Y.H. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Point mutation in a virus-like capsid drives symmetry reduction to form tetrahedral cages.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8V4Q
 
 | | Myxococcus xanthus EncA 3xHis pore mutant with tetrahedral symmetry | | Descriptor: | Type 1 encapsulin shell protein EncA | | Authors: | Szyszka, T.N, Andreas, M.P, Lie, F, Miller, L.M, Adamson, L.S.R, Fatehi, F, Twarock, R, Draper, B.E, Jarrold, M.F, Giessen, T.W, Lau, Y.H. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Point mutation in a virus-like capsid drives symmetry reduction to form tetrahedral cages.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1HJ8
 
 | | 1.00 AA Trypsin from Atlantic Salmon | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Leiros, H.-K.S, Mcsweeney, S.M, Smalas, A.O. | | Deposit date: | 2001-01-09 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic Resolution Structure of Trypsin Provide Insight Into Structural Radiation Damage
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IA7
 
 | | CRYSTAL STRUCTURE OF THE CELLULASE CEL9M OF C. CELLULOLYTICIUM IN COMPLEX WITH CELLOBIOSE | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULASE CEL9M, ... | | Authors: | Parsiegla, G, Belaich, A, Belaich, J.P, Haser, R. | | Deposit date: | 2001-03-22 | | Release date: | 2002-10-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the cellulase Cel9M enlightens structure/function relationships of the variable catalytic modules in glycoside hydrolases.
Biochemistry, 41, 2002
|
|
7VCJ
 
 | | Arginine kinase H227A from Daphnia magna | | Descriptor: | Arginine kinase, NITRATE ION, PHOSPHATE ION | | Authors: | Kim, D.S, Jang, K, Kim, W.S, Kim, Y.J, Park, J.H. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of H227A Mutant of Arginine Kinase in Daphnia magna Suggests the Importance of Its Stability.
Molecules, 27, 2022
|
|
