8UUU
 
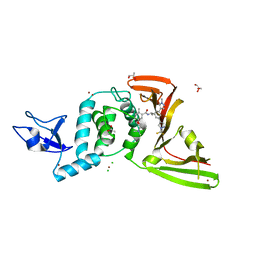 | | SARS-Cov-2 papain-like protease (PLpro) with inhibitor Jun12162 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8ONN
 
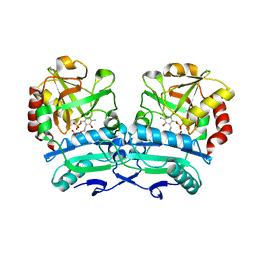 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with 3-aminooxypropionic acid | | Descriptor: | 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
7N46
 
 | |
7VJZ
 
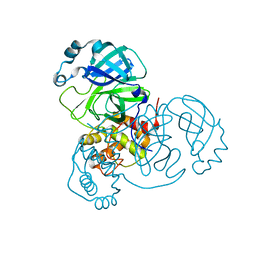 | | Crystal Structure of SARS-CoV-2 Mpro at 1.90 A resolution-7 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Tokay, N. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
8V9H
 
 | | GES-5-NA-1-157 complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Restricted Rotational Flexibility of the C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the GES-5 Carbapenemase.
Acs Infect Dis., 10, 2024
|
|
7ONM
 
 | | Carbonic anhydrase II mutant (N67G-E69R-I91C) dually binding an IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(4-azanyl-9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
7VK7
 
 | | Crystal Structure of SARS-CoV-2 Mpro at 2.4 A resolution-11 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Dag, C. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
7ONQ
 
 | | Carbonic anhydrase II mutant (E69C) dually binding an IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(4-azanyl-9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
7NE1
 
 | | Structure of the complex between Netrin-1 and its receptor Neogenin | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Robinson, R.A, Griffiths, S.C, van de Haar, L.L, Malinauskas, T, van Battum, E.Y, Zelina, P, Schwab, R.A, Karia, D, Malinauskaite, L, Brignani, S, van den Munkhof, M, Dudukcu, O, De Ruiter, A.A, Van den Heuvel, D.M.A, Bishop, B, Elegheert, J, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Simultaneous binding of Guidance Cues NET1 and RGM blocks extracellular NEO1 signaling.
Cell, 184, 2021
|
|
7VJX
 
 | | Crystal Structure of SARS-CoV-2 Mpro at 2.20 A resolution-12 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Usta, G. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
6MPT
 
 | | TagT bound to LI-WTA | | Descriptor: | 2-(acetylamino)-2-deoxy-1-O-[(S)-{[(R)-{[(2Z,6Z,10E,14E,18E)-3,7,11,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaen-1-yl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-alpha-D-glucopyranose, CHLORIDE ION, Polyisoprenyl-teichoic acid--peptidoglycan teichoic acid transferase TagT | | Authors: | Owens, T.W, Schaefer, K, Kahne, D, Walker, S. | | Deposit date: | 2018-10-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Substrate Preferences Establish the Order of Cell Wall Assembly in Staphylococcus aureus.
J. Am. Chem. Soc., 140, 2018
|
|
7ONV
 
 | | Carbonic anhydrase II mutant (I91C) dually binding an IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(4-azanyl-9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-26 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
5TTA
 
 | | A 1.85A X-Ray Structure from Peptoclostridium difficile 630 of a Hypothetical Protein | | Descriptor: | Putative exported protein | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Cordona-Correa, A, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-02 | | Release date: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A 1.85A X-Ray Structure from Peptoclostridium difficile 630 of a Hypothetical Protein
To Be Published
|
|
8U8M
 
 | | X-ray crystal structure of TEBP-1 MCD2 homodimer | | Descriptor: | COBALT (II) ION, Double-strand telomeric DNA-binding proteins 1 | | Authors: | Nandakumar, J, Padmanaban, S. | | Deposit date: | 2023-09-18 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Caenorhabditis elegans telomere-binding proteins TEBP-1 and TEBP-2 adapt the Myb module to dimerize and bind telomeric DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7VK4
 
 | |
5C82
 
 | |
8UKT
 
 | | RNA polymerase II elongation complex with Fapy-dG lesion with AMP added | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Hou, P, Oh, J, Wang, D. | | Deposit date: | 2023-10-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Molecular Mechanism of RNA Polymerase II Transcriptional Mutagenesis by the Epimerizable DNA Lesion, Fapy·dG.
J.Am.Chem.Soc., 146, 2024
|
|
7VK2
 
 | | Crystal Structure of SARS-CoV-2 Mpro at 2.0 A resolution -9 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Gul, M. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
5TOI
 
 | |
7VK8
 
 | | Crystal structure of SARS-CoV-2 Mpro at 2.4 A Resolution | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Usta, G. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
8OE5
 
 | |
8RV6
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 2 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-(4-hydroxyphenyl)benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
5TOT
 
 | |
8UUF
 
 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun11941 | | Descriptor: | ACETATE ION, CHLORIDE ION, N-{(1R)-1-[(3M,5P)-3,5-bis(1-methyl-1H-pyrazol-4-yl)phenyl]ethyl}-5-[2-(dimethylamino)ethoxy]-2-methylbenzamide, ... | | Authors: | Ansari, A, Tan, B, Riuz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
7NE0
 
 | | Structure of the ternary complex between Netrin-1, Repulsive-Guidance Molecule-B (RGMB) and Neogenin | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Robinson, R.A, Griffiths, S.C, van de Haar, L.L, Malinauskas, T, van Battum, E.Y, Zelina, P, Schwab, R.A, Karia, D, Malinauskaite, L, Brignani, S, van den Munkhof, M, Dudukcu, O, De Ruiter, A.A, Van den Heuvel, D.M.A, Bishop, B, Elegheert, J, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Simultaneous binding of Guidance Cues NET1 and RGM blocks extracellular NEO1 signaling.
Cell, 184, 2021
|
|
