3L9Q
 
 | |
6IHG
 
 | | N terminal domain of Mycobacterium avium complex Lon protease | | Descriptor: | Lon protease | | Authors: | Chen, X.Y, Zhang, S.J, Bi, F.K, Guo, C.Y, Yao, H.W, Lin, D.H. | | Deposit date: | 2018-09-29 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Crystal structure of the N domain of Lon protease from Mycobacterium avium complex.
Protein Sci., 28, 2019
|
|
7LMW
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-((4-(4-carboxyphenoxy)benzyl)oxy)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 7-methyl-3-(1~{H}-pyrazol-4-yl)-1~{H}-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2021-02-06 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
7LML
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-(((3-(4-Carboxyphenoxy)benzyl)oxy)methyl)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 6-iodanyl-1~{H}-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
4AGO
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5174 | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, TERT-BUTYL [3-(3-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4-HYDROXY-5-IODOPHENYL)PROP-2-YN-1-YL]CARBAMATE, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Fersht, A.R, Boeckler, F.M. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
4AGN
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5116 | | Descriptor: | 2-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4-(3-HYDROXYPROP-1-YN-1-YL)-6-IODOPHENOL, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Boeckler, F.M, Fersht, A.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
4AGQ
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5196 | | Descriptor: | 2-{[4-(diethylamino)piperidin-1-yl]methyl}-6-iodo-4-[3-(phenylamino)prop-1-yn-1-yl]phenol, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Boeckler, F.M, Fersht, A.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
4AGP
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5176 | | Descriptor: | 2-{[4-(diethylamino)piperidin-1-yl]methyl}-6-iodo-4-(3-phenoxyprop-1-yn-1-yl)phenol, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Fersht, A.R, Boeckler, F.M. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
4AGL
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan784 | | Descriptor: | 2,4-BIS(IODANYL)-6-[[METHYL-(1-METHYLPIPERIDIN-4-YL)AMINO]METHYL]PHENOL, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Boeckler, F.M, Fersht, A.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
4AGM
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5086 | | Descriptor: | 2-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4,6-DIIODOPHENOL, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Boeckler, F.M, Fersht, A.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
5EL2
 
 | |
3TF2
 
 | | Crystal structure of the cap free human translation initiation factor eIF4E | | Descriptor: | Eukaryotic translation initiation factor 4E, UNKNOWN ATOM OR ION | | Authors: | Siddiqui, N, Tempel, W, Nedyalkova, L, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Borden, K.L.B, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-08-15 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the allosteric effects of 4EBP1 on the eukaryotic translation initiation factor eIF4E.
J.Mol.Biol., 415, 2012
|
|
3U7X
 
 | | Crystal structure of the human eIF4E-4EBP1 peptide complex without cap | | Descriptor: | Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E-binding protein 1, SULFATE ION, ... | | Authors: | Siddiqui, N, Tempel, W, Nedyalkova, L, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Borden, K.L.B, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-14 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into the Allosteric Effects of 4EBP1 on the Eukaryotic Translation Initiation Factor eIF4E.
J.Mol.Biol., 415, 2012
|
|
3RKF
 
 | | Crystal structure of guanine riboswitch C61U/G37A double mutant bound to thio-guanine | | Descriptor: | 2-amino-1,9-dihydro-6H-purine-6-thione, COBALT HEXAMMINE(III), Guanine riboswitch | | Authors: | Buck, J, Wacker, A, Warkentin, E, Woehnert, J, Wirmer-Bartoschek, J, Schwalbe, H. | | Deposit date: | 2011-04-18 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Influence of ground-state structure and Mg2+ binding on folding kinetics of the guanine-sensing riboswitch aptamer domain.
Nucleic Acids Res., 39, 2011
|
|
5K79
 
 | | Structure and anti-HIV activity of CYT-CVNH, a new cyanovirin-n homolog | | Descriptor: | 1,2-ETHANEDIOL, Cyanovirin-N domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Matei, E, Basu, R, Furey, W, Shi, J, Calnan, C, Aiken, C, Gronenborn, A.M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Glycan Binding of a New Cyanovirin-N Homolog.
J.Biol.Chem., 291, 2016
|
|
6VBK
 
 | | Crystal structure of N-terminal domain of Mycobacterium tuberculosis complex Lon protease | | Descriptor: | GLYCEROL, Lon211 | | Authors: | Bi, F.K, Chen, C, Chen, X.Y, Guo, C.Y, Lin, D.H. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the N domain of Lon protease from Mycobacterium avium complex.
Protein Sci., 28, 2019
|
|
2Y6X
 
 | | Structure of Psb27 from Thermosynechococcus elongatus | | Descriptor: | CHLORIDE ION, PHOTOSYSTEM II 11 KD PROTEIN | | Authors: | Michoux, F, Takasaka, K, Boehm, M, Nixon, P.J, Murray, J.W. | | Deposit date: | 2011-01-27 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Psb27 Assembly Factor at 1.6A: Implications for Binding to Photosystem II.
Photosynth.Res., 110, 2012
|
|
3ZQ8
 
 | |
3EV3
 
 | | Crystal Structure of Ribonuclease A in 70% t-Butanol | | Descriptor: | Ribonuclease pancreatic, TERTIARY-BUTYL ALCOHOL | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EUY
 
 | | Crystal Structure of Ribonuclease A in 50% Dioxane | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EV6
 
 | | Crystal Structure of Ribonuclease A in 50% R,S,R-Bisfuranol | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-ol, Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EUX
 
 | | Crystal Structure of Crosslinked Ribonuclease A | | Descriptor: | Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EV4
 
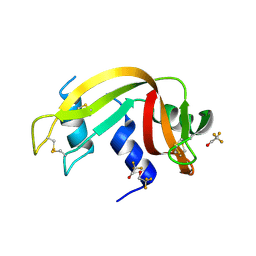 | | Crystal Structure of Ribonuclease A in 50% Trifluoroethanol | | Descriptor: | Ribonuclease pancreatic, TRIFLUOROETHANOL | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EZM
 
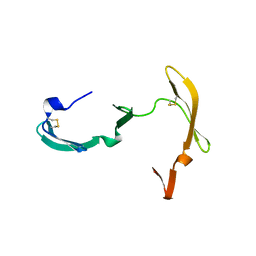 | | CYANOVIRIN-N | | Descriptor: | PROTEIN (CYANOVIRIN-N) | | Authors: | Yang, F, Wlodawer, A. | | Deposit date: | 1998-12-15 | | Release date: | 1998-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of cyanovirin-N, a potent HIV-inactivating protein, shows unexpected domain swapping.
J.Mol.Biol., 288, 1999
|
|
3EUZ
 
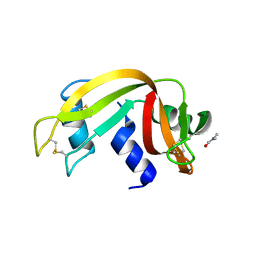 | | Crystal Structure of Ribonuclease A in 50% Dimethylformamide | | Descriptor: | DIMETHYLFORMAMIDE, Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
