8RV8
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 5 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]-2-chloranyl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
7OWO
 
 | | HsNMT1 in complex with both MyrCoA and N-acetylated KSFSKPR peptide | | Descriptor: | COENZYME A, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2021-06-18 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
5U16
 
 | | Structure of human MR1-2-OH-1-NA in complex with human MAIT A-F7 TCR | | Descriptor: | 2-hydroxynaphthalene-1-carbaldehyde, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Keller, A.N, Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2016-11-27 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Drugs and drug-like molecules can modulate the function of mucosal-associated invariant T cells.
Nat. Immunol., 18, 2017
|
|
7OWR
 
 | | HsNMT1 in complex with both MyrCoA and peptide GGKSFSKPR | | Descriptor: | GLY-GLY-LYS-SER-PHE-SER-LYS-PRO-ARG, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2021-06-18 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
7V6I
 
 | |
7NBD
 
 | | Crystal structure of human serine racemase in complex with DSiP fragment Z235449082, XChem fragment screen. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Koulouris, C.R, Roe, S.M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.865 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
5UG2
 
 | | CcP gateless cavity | | Descriptor: | 6-fluoro-2-methylimidazo[1,2-a]pyridin-3-amine, PROTOPORPHYRIN IX CONTAINING FE, Peroxidase | | Authors: | Stein, R.M, Fischer, M, Shoichet, B.K. | | Deposit date: | 2017-01-06 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Testing inhomogeneous solvation theory in structure-based ligand discovery.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6MM2
 
 | |
8UQW
 
 | | Round 18 Arylesterase Variant of Apo-Phosphotriesterase Measured at 13 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphotriesterase variant PTE-R18 | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RV9
 
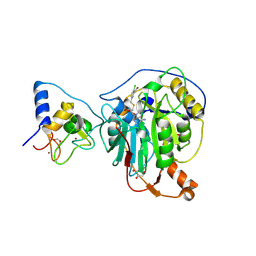 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 6 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-2-chloranyl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
5CNQ
 
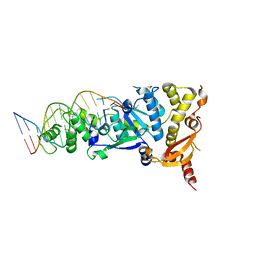 | | Crystal structure of the Holliday junction-resolving enzyme GEN1 (WT) in complex with product DNA, Mg2+ and Mn2+ ions | | Descriptor: | DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MANGANESE (II) ION, Nuclease-like protein, ... | | Authors: | Liu, Y.J, Freeman, A.D.J, Declais, A.C, Wilson, T.J, Gartner, A, Lilley, D.M.J. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Crystal Structure of a Eukaryotic GEN1 Resolving Enzyme Bound to DNA.
Cell Rep, 13, 2015
|
|
7NBY
 
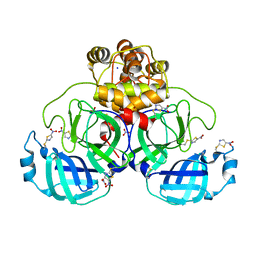 | | Crystal structure of SU3327 (halicin) covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | 5-nitro-1,3-thiazole, CHLORIDE ION, Main Protease, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of SU3327 (halicin) covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2.
To Be Published
|
|
5U2M
 
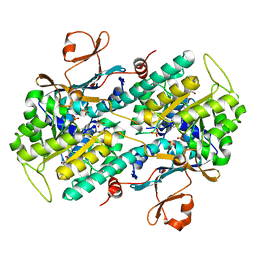 | | Crystal structure of human NAMPT with A-1293201 | | Descriptor: | N-[4-({[(3S)-oxolan-3-yl]methyl}carbamoyl)phenyl]-1,3-dihydro-2H-isoindole-2-carboxamide, Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Longenecker, K.L, Raich, D, Korepanova, A.V. | | Deposit date: | 2016-11-30 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Discovery and Characterization of Novel Nonsubstrate and Substrate NAMPT Inhibitors.
Mol. Cancer Ther., 16, 2017
|
|
5CO3
 
 | |
8UUG
 
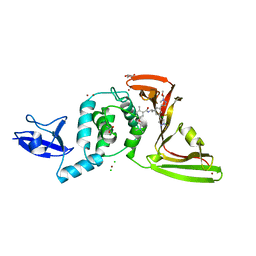 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun12303 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
7VWG
 
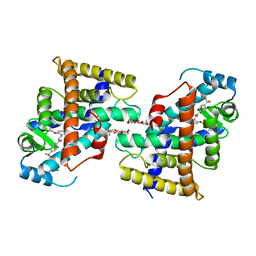 | |
7P4J
 
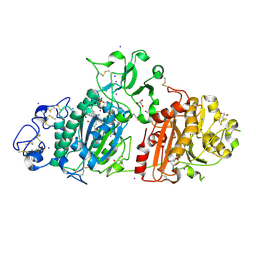 | | Crystal structure of Autotaxin and tetrahydrocannabinol | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A, Hausmann, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Linking medicinal cannabis to autotaxin-lysophosphatidic acid signaling.
Life Sci Alliance, 6, 2023
|
|
8RZC
 
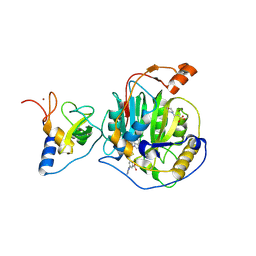 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 11 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-imidazol-1-yl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8UUH
 
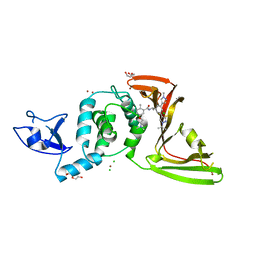 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun12199 | | Descriptor: | 5-[2-(dimethylamino)ethoxy]-2-methyl-N-[(1R)-1-{(3M,5P)-3-(1-methyl-1H-pyrazol-4-yl)-5-[1-(propan-2-yl)-1H-pyrazol-4-yl]phenyl}ethyl]benzamide, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
7NBC
 
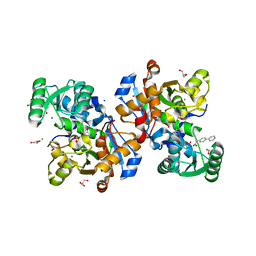 | | Crystal structure of human serine racemase in complex with DSiP fragment Z2856434779, XChem fragment screen. | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, ... | | Authors: | Koulouris, C.R, Roe, S.M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
7VC7
 
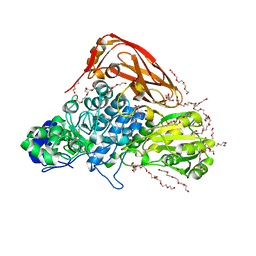 | | The structure of beta-xylosidase from Phanerochaete chrysosporium(PcBxl3) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kojima, K, Sunagawa, N, Igarashi, K. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Comparison of glycoside hydrolase family 3 beta-xylosidases from basidiomycetes and ascomycetes reveals evolutionarily distinct xylan degradation systems.
J.Biol.Chem., 298, 2022
|
|
8UUW
 
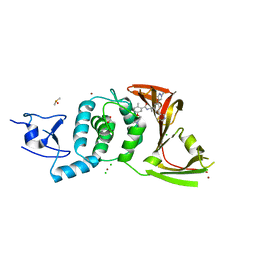 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun12145 | | Descriptor: | 5-[2-(dimethylamino)ethoxy]-2-methyl-N-{(1R)-1-[(3P,5M)-3-(1-methyl-1H-pyrazol-4-yl)-5-(1,3-thiazol-5-yl)phenyl]ethyl}benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8V33
 
 | |
7NBF
 
 | | Crystal structure of human serine racemase in complex with DSiP fragment Z126932614, XChem fragment screen. | | Descriptor: | 1,2-ETHANEDIOL, 2-[(methylsulfonyl)methyl]-1H-benzimidazole, CALCIUM ION, ... | | Authors: | Koulouris, C.R, Roe, S.M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
8V34
 
 | |
