3L4A
 
 | |
3KL2
 
 | | Crystal structure of a putative isochorismatase from Streptomyces avermitilis | | Descriptor: | Putative isochorismatase, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-06 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative isochorismatase from Streptomyces avermitilis
To be Published
|
|
3L3C
 
 | | Crystal structure of the Bacillus anthracis glmS ribozyme bound to Glc6P | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-12-16 | | Release date: | 2009-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
3BS2
 
 | | Crystal Structure of Monomine | | Descriptor: | GLYCEROL, Lipocalin | | Authors: | Mans, B.J, Ribeiro, J.M, Andersen, J.F. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure, function, and evolution of biogenic amine-binding proteins in soft ticks.
J.Biol.Chem., 283, 2008
|
|
3L4I
 
 | | CRYSTAL STRUCTURE OF THE N-TERMINAL DOMAIN OF HSP70 (CGD2_20) FROM Cryptosporidium PARVUM IN COMPLEX WITH ADP and inorganic phosphate | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Pizarro, J.C, Wernimont, A.K, Hutchinson, A, Sullivan, H, Lew, J, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-20 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CRYSTAL STRUCTURE OF THE N-TERMINAL DOMAIN OF HSP70 (CGD2_20) FROM Cryptosporidium PARVUM IN COMPLEX WITH ADP and inorganic phosphate
To be Published
|
|
3L4Y
 
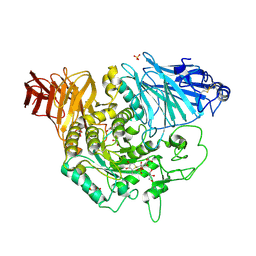 | | Crystal complex of N-terminal Human Maltase-Glucoamylase with NR4-8II | | Descriptor: | (1S,2R,3S,4R)-1-{(1S)-2-[(2R,3S,4S)-3,4-dihydroxy-2-(hydroxymethyl)tetrahydrothiophenium-1-yl]-1-hydroxyethyl}-2,3,4,5-tetrahydroxypentyl sulfate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New glucosidase inhibitors from an ayurvedic herbal treatment for type 2 diabetes: structures and inhibition of human intestinal maltase-glucoamylase with compounds from Salacia reticulata.
Biochemistry, 49, 2010
|
|
3KMX
 
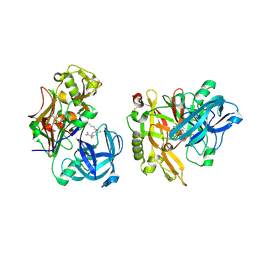 | | Structure of BACE bound to SCH346572 | | Descriptor: | 4-butoxy-3-chlorobenzyl imidothiocarbamate, Beta-secretase 1 | | Authors: | Strickland, C, Wang, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3KL8
 
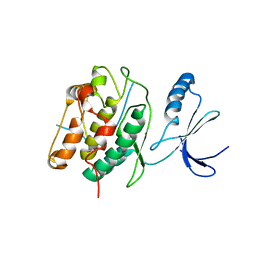 | | CaMKIINtide Inhibitor Complex | | Descriptor: | Calcium/calmodulin dependent protein kinase II, Calcium/calmodulin-dependent protein kinase II inhibitor 1 | | Authors: | Kuriyan, J, Chao, L.H, Pellicena, P, Deindl, S, Barclay, L.A, Schulman, H. | | Deposit date: | 2009-11-06 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.372 Å) | | Cite: | Intersubunit capture of regulatory segments is a component of cooperative CaMKII activation.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3BUQ
 
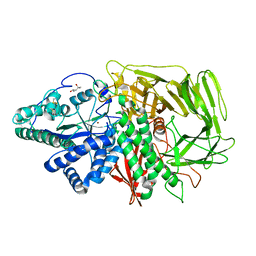 | |
3KLQ
 
 | | Crystal Structure of the Minor Pilin FctB from Streptococcus pyogenes 90/306S | | Descriptor: | GLYCEROL, Putative pilus anchoring protein | | Authors: | Linke, C, Young, P.G, Bunker, R.D, Caradoc-Davies, T.T, Baker, E.N. | | Deposit date: | 2009-11-08 | | Release date: | 2010-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the minor pilin FctB reveals determinants of Group A streptococcal pilus anchoring
J.Biol.Chem., 285, 2010
|
|
3BXB
 
 | |
3L0M
 
 | |
3KN0
 
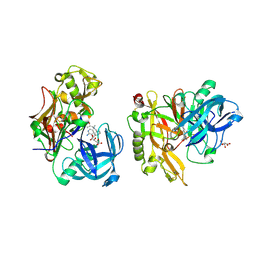 | | Structure of BACE bound to SCH708236 | | Descriptor: | 3-[2-(3-{[(furan-2-ylmethyl)(methyl)amino]methyl}phenyl)ethyl]pyridin-2-amine, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Wang, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3C3L
 
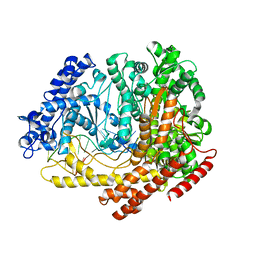 | |
3KQM
 
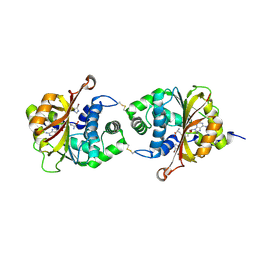 | | Crystal Structure of hPNMT in Complex AdoHcy and 4-Bromo-1H-imidazole | | Descriptor: | 4-bromo-1H-imidazole, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3KQS
 
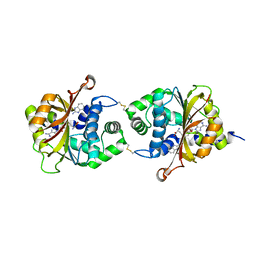 | | Crystal Structure of hPNMT in Complex AdoHcy and 2-Aminobenzimidazole | | Descriptor: | 1H-benzimidazol-2-amine, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3C8M
 
 | |
3C5G
 
 | | Structure of a ternary complex of the R517K Pol lambda mutant | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*DCP*DAP*DGP*DTP*DAP*(2DT))-3'), ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Foley, M.C, Pedersen, L.C, Schlick, T, Kunkel, T.A. | | Deposit date: | 2008-01-31 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate-induced DNA strand misalignment during catalytic cycling by DNA polymerase lambda.
Embo Rep., 9, 2008
|
|
3L4L
 
 | |
3KUX
 
 | | Structure of the YPO2259 putative oxidoreductase from Yersinia pestis | | Descriptor: | CHLORIDE ION, Putative oxidoreductase | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Kwon, K, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the YPO2259 putative oxidoreductase from Yersinia pestis
To be Published
|
|
3CCL
 
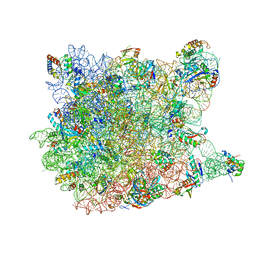 | |
3L4W
 
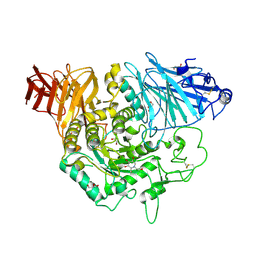 | | Crystal complex of N-terminal Human Maltase-Glucoamylase with miglitol | | Descriptor: | (2R,3R,4R,5S)-1-(2-hydroxyethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New glucosidase inhibitors from an ayurvedic herbal treatment for type 2 diabetes: structures and inhibition of human intestinal maltase-glucoamylase with compounds from Salacia reticulata.
Biochemistry, 49, 2010
|
|
3KXY
 
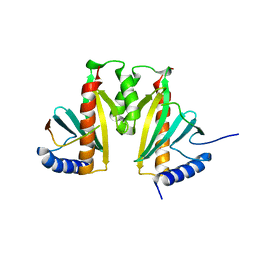 | | Crystal Structure of the ExsC-ExsE Complex | | Descriptor: | Exoenzyme S synthesis protein C, ExsE | | Authors: | Vogelaar, N.J, Robinson, H.H, Schubot, F.D. | | Deposit date: | 2009-12-04 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Analysis of the Crystal Structure of the ExsC.ExsE Complex Reveals Distinctive Binding Interactions of the Pseudomonas aeruginosa Type III Secretion Chaperone ExsC with ExsE and ExsD.
Biochemistry, 49, 2010
|
|
3C7K
 
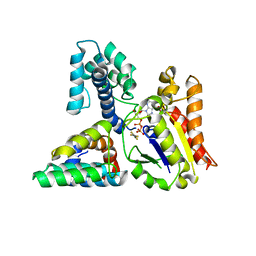 | | Molecular architecture of Galphao and the structural basis for RGS16-mediated deactivation | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(o) subunit alpha, MAGNESIUM ION, ... | | Authors: | Slep, K.C, Kercher, M.A, Wieland, T, Chen, C, Simon, M.I, Sigler, P.B. | | Deposit date: | 2008-02-07 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular architecture of G{alpha}o and the structural basis for RGS16-mediated deactivation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3KPJ
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and Bound Phosphate | | Descriptor: | PHOSPHATE ION, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-16 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
