8PWD
 
 | | Crystal structure of VDR in complex Des-C-Ring and Aromatic-D-Ring analog 3c | | Descriptor: | (1R,3S,5Z)-4-methylidene-5-[(E)-3-[3-[7,7,7-tris(fluoranyl)-6-oxidanyl-6-(trifluoromethyl)hept-3-ynyl]phenyl]pent-2-enylidene]cyclohexane-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2023-07-20 | | Release date: | 2024-08-14 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Further Studies on the Highly Active Des-C-Ring and Aromatic-D-Ring Analogues of 1 alpha ,25-Dihydroxyvitamin D 3 (Calcitriol): Refinement of the Side Chain.
J.Med.Chem., 66, 2023
|
|
8PWF
 
 | | crystal structure of VDR in complex with Des-C-Ring and Aromatic-D-Ring analog 2 | | Descriptor: | (1R,3S,5Z)-4-methylidene-5-[(E)-3-[3-[7,7,7-tris(fluoranyl)-6-oxidanyl-6-(trifluoromethyl)heptyl]phenyl]pent-2-enylidene]cyclohexane-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | rochel, N. | | Deposit date: | 2023-07-20 | | Release date: | 2024-08-14 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Further Studies on the Highly Active Des-C-Ring and Aromatic-D-Ring Analogues of 1 alpha ,25-Dihydroxyvitamin D 3 (Calcitriol): Refinement of the Side Chain.
J.Med.Chem., 66, 2023
|
|
8HAV
 
 | | An auto-activation mechanism of plant non-specific phospholipase C | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Non-specific phospholipase C4 | | Authors: | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
6XX2
 
 | |
8YMI
 
 | | BRD4-BD1 in complex with 2-{[(3S)-1-benzylpyrrolidin-3-yl]methyl}-5-methyl-7-(1-methylpyrazol-3-yl)pyrazolo[4,3-c]pyridin-4-one | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(3S)-1-benzylpyrrolidin-3-yl]methyl}-5-methyl-7-(1-methylpyrazol-3-yl)pyrazolo[4,3-c]pyridin-4-one, Bromodomain-containing protein 4 | | Authors: | Sasaki, C, Miyaguchi, I, Hagihara, S, Ishizawa, K, Endo, J. | | Deposit date: | 2024-03-09 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Discovery of a potent, orally available pyrazolopyridone derivative as a novel selective BET BD1 Inhibitor (Part 2).
To Be Published
|
|
8YMG
 
 | | BRD4-BD1 in complex with 7-[4-chloro-1-(tetrahydropyran-4-ylmethyl)imidazol-2-yl]-5-methyl-2-{[(2R)-2-methyl-4-methylsulfonyl-piperazin-1-yl]methyl}furo[3,2-c]pyridin-4-one | | Descriptor: | 7-[4-chloro-1-(tetrahydropyran-4-ylmethyl)imidazol-2-yl]-5-methyl-2-{[(2R)-2-methyl-4-methylsulfonyl-piperazin-1-yl]methyl}furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4 | | Authors: | Sasaki, C, Miyaguchi, I, Hagihara, S, Ishizawa, K, Endo, J. | | Deposit date: | 2024-03-09 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.007 Å) | | Cite: | Discovery of a potent, orally available furopyridine derivative as a novel selective BET BD1 Inhibitor (Part 1).
To Be Published
|
|
8YMF
 
 | | BRD4-BD1 in complex with 7-[2-(cyclopropylmethoxy)phenyl]-5-methyl-2-[(1-methylsulfonyl-4-piperidyl)methyl]pyrazolo[4,3-c]pyridin-4-one | | Descriptor: | 7-[2-(cyclopropylmethoxy)phenyl]-5-methyl-2-[(1-methylsulfonylpiperidin-4-yl)methyl]pyrazolo[4,3-c]pyridin-4-one, Bromodomain-containing protein 4 | | Authors: | Sasaki, C, Miyaguchi, I, Hagihara, S, Ishizawa, K, Endo, J. | | Deposit date: | 2024-03-09 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.015 Å) | | Cite: | Discovery of a potent, orally available pyrazolopyridone derivative as a novel selective BET BD1 Inhibitor (Part 2).
To Be Published
|
|
8YMH
 
 | | BRD4-BD1 in complex with 5-methyl-2-{[(2R)-2-methyl-4-methylsulfonyl-piperazin-1-yl]methyl}-7-(1-methylpyrazol-3-yl)furo[3,2-c]pyridin-4-one | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-2-{[(2R)-2-methyl-4-methylsulfonyl-piperazin-1-yl]methyl}-7-(1-methylpyrazol-3-yl)furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4 | | Authors: | Sasaki, C, Miyaguchi, I, Hagihara, S, Ishizawa, K, Endo, J. | | Deposit date: | 2024-03-09 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Discovery of a potent, orally available furopyridine derivative as a novel selective BET BD1 Inhibitor (Part 1).
To Be Published
|
|
8R5C
 
 | | Crystal structure of human TRIM7 PRYSPRY domain bound to (2-(1-oxoisoindolin-2-yl)-3-phenylpropanoyl)-L-glutamine | | Descriptor: | (2~{S})-5-azanyl-5-oxidanylidene-2-[[(2~{S})-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)-3-phenyl-propanoyl]amino]pentanoic acid, 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM7, ... | | Authors: | Munoz Sosa, C.J, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A C-Degron Structure-Based Approach for the Development of Ligands Targeting the E3 Ligase TRIM7.
Acs Chem.Biol., 19, 2024
|
|
6XX3
 
 | |
8YME
 
 | | BRD4-BD1 in complex with N-cyclohexyl-1-methyl-6-{[5-methyl-7-(1-methylpyrazol-3-yl)-4-oxo-pyrazolo[4,3-c]pyridin-2-yl]methyl}azepan-1-ium-4-carboxamide 2,2,2-trifluoroacetate | | Descriptor: | (4R,6S)-N-cyclohexyl-1-methyl-6-[[5-methyl-7-(1-methylpyrazol-3-yl)-4-oxidanylidene-pyrazolo[4,3-c]pyridin-2-yl]methyl]azepane-4-carboxamide, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Sasaki, C, Miyaguchi, I, Hagihara, S, Ishizawa, K, Endo, J. | | Deposit date: | 2024-03-09 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Discovery of a potent, orally available pyrazolopyridone derivative as a novel selective BET BD1 Inhibitor (Part 2).
To Be Published
|
|
6Y38
 
 | | Crystal structure of Whirlin PDZ3 in complex with Myosin 15a C-terminal PDZ binding motif peptide | | Descriptor: | Chains: C,D, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-02-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
8T3J
 
 | | Crystal structure of native exfoliative toxin C (ExhC) from Mammaliicoccus sciuri | | Descriptor: | Exfoliative toxin C | | Authors: | Calil, F.A, Gismene, C, Hernandez Gonzalez, J.E, Ziem Nascimento, A.F, Santisteban, A.R.N, Arni, R.K, Barros Mariutti, R. | | Deposit date: | 2023-06-07 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Necrotic activity of ExhC from Mammaliicoccus sciuri is mediated by specific amino acid residues.
Int.J.Biol.Macromol., 254, 2024
|
|
7ZC2
 
 | | Dipeptide and tripeptide Permease C (DtpC) | | Descriptor: | Amino acid/peptide transporter | | Authors: | Killer, M, Finocchio, G, Pardon, E, Steyaert, J, Loew, C. | | Deposit date: | 2022-03-25 | | Release date: | 2022-07-06 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM Structure of an Atypical Proton-Coupled Peptide Transporter: Di- and Tripeptide Permease C.
Front Mol Biosci, 9, 2022
|
|
7PPE
 
 | | CRYSTAL STRUCTURE OF NAMPT IN COMPLEX WITH COMPOUND 1 | | Descriptor: | GLYCEROL, N-[4-[(4R)-4-methyl-1-(oxan-4-yl)-6-oxidanylidene-4,5-dihydropyridazin-3-yl]phenyl]-1,3-dihydropyrrolo[3,4-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Hillig, R.C. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Novel NAMPT Inhibitor-Based Antibody-Drug Conjugate Payload Class for Cancer Therapy.
Bioconjug.Chem., 33, 2022
|
|
6OB1
 
 | | Structure of WHB in complex with Ubiquitin Variant | | Descriptor: | Anaphase-promoting complex subunit 2, Ubiquitin | | Authors: | Edmond, R.W, Grace, C.R. | | Deposit date: | 2019-03-19 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein engineering of a ubiquitin-variant inhibitor of APC/C identifies a cryptic K48 ubiquitin chain binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8H28
 
 | | Crystal structure of the K87V mutant of cytochrome c' from Shewanella benthica DB6705 | | Descriptor: | Class II cytochrome c, HEME C | | Authors: | Fujii, S, Sakaguchi, R, Oki, H, Kawahara, K, Ohkubo, T, Fujiyoshi, S, Sambongi, Y. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Contribution of a surface salt bridge to the protein stability of deep-sea Shewanella benthica cytochrome c'.
J.Struct.Biol., 215, 2023
|
|
9CUV
 
 | |
6NZ9
 
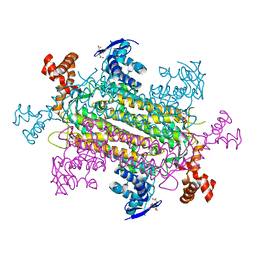 | | Crystal structure of E. coli fumarase C bound to citrate at 1.53 angstrom resolution | | Descriptor: | CITRIC ACID, Fumarate hydratase class II | | Authors: | Stuttgen, G.M, May, J.F, Bhattcharyya, B, Weaver, T.M. | | Deposit date: | 2019-02-13 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Closed fumarase C active-site structures reveal SS Loop residue contribution in catalysis.
Febs Lett., 594, 2020
|
|
7TFS
 
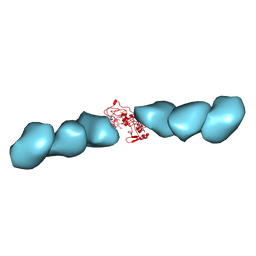 | | Cryo-EM of the OmcE nanowires from Geobacter sulfurreducens | | Descriptor: | Cytochrome c, HEME C | | Authors: | Wang, F, Mustafa, K, Chan, C.H, Joshi, K, Hochbaum, A.I, Bond, D.R, Egelman, E.H. | | Deposit date: | 2022-01-07 | | Release date: | 2022-05-04 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing.
Nat Microbiol, 7, 2022
|
|
5DQN
 
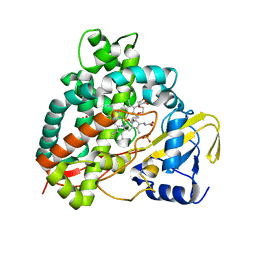 | | Polyethylene 600-bound form of P450 CYP125A3 mutant from Myobacterium Smegmatis - W83Y | | Descriptor: | CITRIC ACID, Cytochrome P450 CYP125, PENTAETHYLENE GLYCOL, ... | | Authors: | Ortiz de Montellano, P.J, Frank, D.J, Waddling, C.A. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.262 Å) | | Cite: | Cytochrome P450 125A4, the Third Cholesterol C-26 Hydroxylase from Mycobacterium smegmatis.
Biochemistry, 54, 2015
|
|
6XX4
 
 | |
8PWM
 
 | | Crystal structure of VDR in complex with Des-C-Ring and Aromatic-D-Ring analog 3b | | Descriptor: | (1R,3S,5Z)-4-methylidene-5-[(E)-3-[3-[7,7,7-tris(fluoranyl)-6-oxidanyl-6-(trifluoromethyl)hept-3-ynyl]phenyl]but-2-enylidene]cyclohexane-1,3-diol, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | Rochel, N. | | Deposit date: | 2023-07-20 | | Release date: | 2024-08-21 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Further Studies on the Highly Active Des-C-Ring and Aromatic-D-Ring Analogues of 1 alpha ,25-Dihydroxyvitamin D 3 (Calcitriol): Refinement of the Side Chain.
J.Med.Chem., 66, 2023
|
|
6HIH
 
 | | Cytochrome c prime beta from Methylococcus capsulatus (Bath) | | Descriptor: | Cytochrome c, GLYCEROL, HEME C, ... | | Authors: | Adams, H, Chicano, T.M, Hough, M.A. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | One fold, two functions: cytochrome P460 and cytochromec'-beta from the methanotrophMethylococcus capsulatus(Bath).
Chem Sci, 10, 2019
|
|
7RXJ
 
 | |
