7YDO
 
 | | Crystal structure of Atg44 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Uncharacterized protein C26A3.14c | | Authors: | Maruyama, T, Noda, N.N. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The mitochondrial intermembrane space protein mitofissin drives mitochondrial fission required for mitophagy.
Mol.Cell, 83, 2023
|
|
7UUB
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI12 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methylidene-L-norvalinamide | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
5T3X
 
 | |
8QPT
 
 | | Crystal structure of pyrophosphatase from Ogataea parapolymorpha | | Descriptor: | GLYCEROL, MAGNESIUM ION, inorganic diphosphatase | | Authors: | Matyuta, I.O, Rodina, E.V, Vorobyeva, N.N, Kurilova, S.A, Bezpalaya, E.Y, Boyko, K.M. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of yeast mitochondrial type pyrophosphatase provides a model to study pathological mutations in its human ortholog.
Biochem.Biophys.Res.Commun., 738, 2024
|
|
6M30
 
 | | Crystal structure of a mutant Staphylococcus equorum manganese superoxide dismutase N73F | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Retnoningrum, D.S, Yoshida, H, Razani, M.D, Meidianto, V.F, Hartanto, A, Artarini, A, Ismaya, W.T. | | Deposit date: | 2020-03-02 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Unprecedented Role of The N73-F124 Pair in The Staphylococcus equorum MnSOD Activity.
Curr Enzym Inhib, 2021
|
|
7MED
 
 | | CDD-1 beta-lactamase in imidazole/MPD 5 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
8TY4
 
 | | MI-30 bound to Mpro of SARS-CoV-2 | | Descriptor: | (1S,3aR,6aS)-2-[(2,4-dichlorophenoxy)acetyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase nsp5 | | Authors: | Blankenship, L.R, Liu, W.R. | | Deposit date: | 2023-08-24 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | MI-30 bound to SARS-CoV-2 Mpro
To Be Published
|
|
7MEG
 
 | | CDD-1 beta-lactamase in imidazole/MPD 30 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
6M0Z
 
 | | X-ray structure of Drosophila dopamine transporter with NET-like mutations (D121G/S426M/F471L) in L-norepinephrine bound form | | Descriptor: | Antibody fragment (Fab) 9D5 Light chain, Antibody fragment (Fab) 9D5 heavy chain, CHLORIDE ION, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-02-24 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
8Q6Q
 
 | |
7UUA
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI8 | | Descriptor: | 3C-like proteinase nsp5, N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
7MEH
 
 | | CDD-1 beta-lactamase in imidazole/MPD 60 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
8TWY
 
 | | Structure of PIK3CA with covalent inhibitor ALO26 | | Descriptor: | 1-(4-{[2-(4-{(4P)-4-[2-amino-4-(difluoromethyl)pyrimidin-5-yl]-6-[(3S)-3-methylmorpholin-4-yl]-1,3,5-triazin-2-yl}piperazin-1-yl)-2-oxoethoxy]methyl}piperidin-1-yl)prop-2-en-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Burke, J.E, Barlow-Busch, I. | | Deposit date: | 2023-08-21 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structure of PIK3CA with covalent inhibitor ALO26
To Be Published
|
|
8Q8N
 
 | | Crystal structure of human GPX4-U46C-I129S-L130S | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Bostock, M.J, Mourao, A, Napolitano, V, Sattler, M, Conrad, M, Popowicz, G. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An ultra-rare variant of GPX4 reveals the structural basis to avert neurodegeneration
To Be Published
|
|
7ONF
 
 | | The binding of p-coumaroyl glucose to glycogen phosphorylase reveals the relationship between structural data and effects on cell metabolome | | Descriptor: | Glycogen phosphorylase, muscle form, INOSINIC ACID, ... | | Authors: | Tsagkarakou, A.S, Koulas, S.M, Kyriakis, E, Drakou, C.E, Leonidas, D.D. | | Deposit date: | 2021-05-25 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure activity relationship of the binding of p-coumaroyl glucose to glycogen phosphorylase and its effect on hepatic cell metabolic pathways
Eur J Med Chem Rep, 3, 2021
|
|
8U05
 
 | |
7MEA
 
 | | CDD-1 beta-lactamase in imidazole/MPD 1 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
8TZI
 
 | | Human equilibrative nucleoside transporter-1, JH-ENT-01 bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[4-[3-[3,4-dimethoxy-5-[[4-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]methoxy]phenyl]carbonyloxypropyl]-1,4-diazepan-1-yl]propyl 3,4,5-trimethoxybenzoate, Equilibrative nucleoside transporter 1 | | Authors: | Wright, N.J, Lee, S.Y. | | Deposit date: | 2023-08-26 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Rational ENT1 inhibition as a pathway for neuropathic pain relief
To Be Published
|
|
7UCX
 
 | | LRP8 11H1 Fab complexed to a cyclized CR1 peptide | | Descriptor: | 11H1 Fab Heavy chain, 11H1 Fab Light chain, Cyclized CR1 peptide, ... | | Authors: | Argiriadi, M.A, Deng, K, Egan, D, Gao, L, Gizatullin, F, Harlan, J, Karaoglu, D, Qiu, W, Goodearl, A. | | Deposit date: | 2022-03-17 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The use of cyclic peptide antigens to generate LRP8 specific antibodies
Front Drug Discov (Lausanne), 2, 2023
|
|
6M0F
 
 | | X-ray structure of Drosophila dopamine transporter with subsiteB mutations (D121G/S426M) in substrate-free form | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Antibody fragment (9D5) Light chain, Antibody fragment (9D5) heavy chain, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-02-21 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
7MEB
 
 | | CDD-1 beta-lactamase in imidazole/MPD 2 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
5TOQ
 
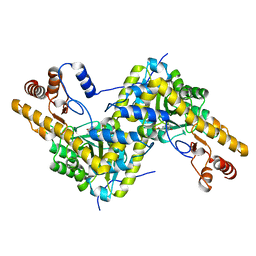 | | High resolution crystal structure of AAT | | Descriptor: | Aspartate aminotransferase, cytoplasmic | | Authors: | Mueser, T.C, Dajnowicz, S, Kovalevsky, A. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Direct evidence that an extended hydrogen-bonding network influences activation of pyridoxal 5'-phosphate in aspartate aminotransferase.
J. Biol. Chem., 292, 2017
|
|
6M2R
 
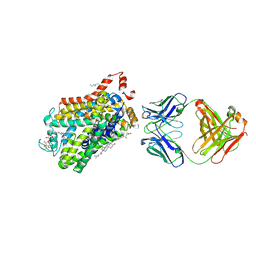 | | X-ray structure of a functional Drosophila dopamine transporter in L-norepinephrine bound form | | Descriptor: | Antibody fragment 9D5 Light chain, Antibody fragment 9D5 heavy chain, CHLORIDE ION, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-02-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
8U0I
 
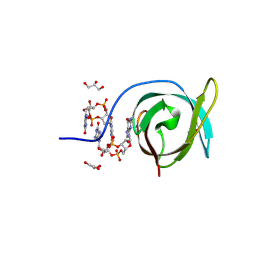 | | Crystal structure of PA0012 complexed with cyclic-di-GMP from Pseudomonas aeruginosa | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), GLYCEROL, PilZ domain-containing protein | | Authors: | Hammons, N.A, Schnicker, N.J, Fuentes, E.J. | | Deposit date: | 2023-08-29 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | Crystal structure of PA0012 complexed with cyclic-di-GMP from Pseudomonas aeruginosa
To Be Published
|
|
7UU9
 
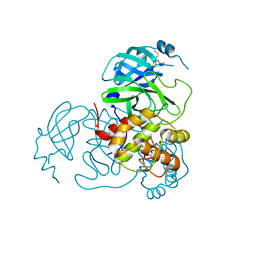 | |
