9GAN
 
 | | CryoEM structure of influenza A RNP-like particle single-stranded assembled with a 12-mer RNA. | | Descriptor: | 2-[3,6-bis(oxidanylidene)-4,5-dihydroxanthen-9-yl]-4-[3-[(2R)-2-oxidanylpropoxy]propylcarbamoyl]benzoic acid, Nucleoprotein, RNA (5'-P(UC)6-FAM3') | | Authors: | Chenavier, F, Ruigrok, R.W.H, Schoehn, G, Ballandras-Colas, A, Crepin, T. | | Deposit date: | 2024-07-29 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Influenza a virus antiparallel helical nucleocapsid-like pseudo-atomic structure.
Nucleic Acids Res., 53, 2025
|
|
9GAT
 
 | | CryoEM structure of the antiparallel double-stranded influenza A RNP-like particle with a 18-mer RNA. | | Descriptor: | 2-[3,6-bis(oxidanylidene)-4,5-dihydroxanthen-9-yl]-4-[3-[(2R)-2-oxidanylpropoxy]propylcarbamoyl]benzoic acid, Nucleoprotein, RNA (5'-P(UC)9-FAM3') | | Authors: | Chenavier, F, Ruigrok, R.W.H, Schoehn, G, Ballandras-Colas, A, Crepin, T. | | Deposit date: | 2024-07-29 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Influenza a virus antiparallel helical nucleocapsid-like pseudo-atomic structure.
Nucleic Acids Res., 53, 2025
|
|
9GAV
 
 | | Focused reconstruction on strand 1 of the influenza A RNP-like particle double-stranded assembled with a 18-mer RNA. | | Descriptor: | 2-[3,6-bis(oxidanylidene)-4,5-dihydroxanthen-9-yl]-4-[3-[(2R)-2-oxidanylpropoxy]propylcarbamoyl]benzoic acid, Nucleoprotein, RNA (5'-P(UC)9-FAM3') | | Authors: | Chenavier, F, Ruigrok, R.W.H, Schoehn, G, Ballandras-Colas, A, Crepin, T. | | Deposit date: | 2024-07-29 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Influenza a virus antiparallel helical nucleocapsid-like pseudo-atomic structure.
Nucleic Acids Res., 53, 2025
|
|
9BZG
 
 | | Targeting N-Myc in Neuroblastoma with Selective Aurora Kinase A Degraders | | Descriptor: | 7-cyclopentyl-N,N-dimethyl-2-[4-(methylcarbamoyl)anilino]-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A | | Authors: | Baker, Z.D, Tang, J, Shi, K, Aihara, H, Levinson, N.M, Harki, D.A. | | Deposit date: | 2024-05-24 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting N-Myc in neuroblastoma with selective Aurora kinase A degraders.
Cell Chem Biol, 32, 2025
|
|
9BZL
 
 | | Targeting N-Myc in Neuroblastoma with Selective Aurora Kinase A Degraders | | Descriptor: | 7-cyclopentyl-N,N-dimethyl-2-{[5-(methylcarbamoyl)pyridin-2-yl]amino}-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A | | Authors: | Baker, Z.D, Tang, J, Shi, K, Aihara, H, Levinson, N.M, Harki, D.A. | | Deposit date: | 2024-05-24 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Targeting N-Myc in neuroblastoma with selective Aurora kinase A degraders.
Cell Chem Biol, 32, 2025
|
|
5E32
 
 | |
5KLC
 
 | | Structure of CBM_E1, a novel carbohydrate-binding module found by sugar cane soil metagenome | | Descriptor: | Carbohydrate binding module E1 | | Authors: | Liberato, M.V, Campos, B.M, Zeri, A.C.M, Squina, F.M. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | A Novel Carbohydrate-binding Module from Sugar Cane Soil Metagenome Featuring Unique Structural and Carbohydrate Affinity Properties.
J.Biol.Chem., 291, 2016
|
|
5KLF
 
 | | Structure of CBM_E1, a novel carbohydrate-binding module found by sugar cane soil metagenome, complexed with cellopentaose and gadolinium ion | | Descriptor: | Carbohydrate binding module E1, GADOLINIUM ATOM, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Liberato, M.V, Campos, B.M, Zeri, A.C.M, Squina, F.M. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | A Novel Carbohydrate-binding Module from Sugar Cane Soil Metagenome Featuring Unique Structural and Carbohydrate Affinity Properties.
J.Biol.Chem., 291, 2016
|
|
8V4U
 
 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | 3C-like proteinase nsp5, N-(methoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-(trifluoromethyl)-L-prolinamide | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | A Second-Generation Oral SARS-CoV-2 Main Protease Inhibitor Clinical Candidate for the Treatment of COVID-19.
J.Med.Chem., 67, 2024
|
|
3DR8
 
 | | Structure of yncA, a putative ACETYLTRANSFERASE from Salmonella typhimurium with its cofactor Acetyl-CoA | | Descriptor: | ACETATE ION, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Singer, A.U, Skarina, T, Onopriyenko, O, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-07-10 | | Release date: | 2008-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of yncA, a putative ACETYLTRANSFERASE from Salmonella typhimurium with its cofactor Acetyl-CoA
To be Published
|
|
6MW0
 
 | | Mle-Phe-Mle-D-Phe. Linear tetrapeptide related to pseudoxylallemycin A. | | Descriptor: | METHANOL, Mle-Phe-Mle-D-Phe Linear tetrapeptide related to pseudoxylallemycin A | | Authors: | Cameron, A.J, Harris, P.W.R, Brimble, M.A, Squire, C.J. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Investigations of the key macrolactamisation step in the synthesis of cyclic tetrapeptide pseudoxylallemycin A.
Org.Biomol.Chem., 17, 2019
|
|
6UR5
 
 | |
7VVR
 
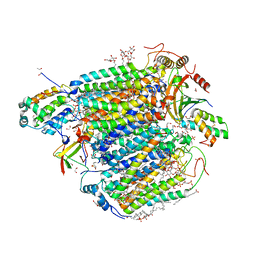 | | Bovine cytochrome c oxidese in CN-bound mixed valence state at 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Tsukihara, T. | | Deposit date: | 2021-11-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic cyanide-probing for cytochrome c oxidase reveals structural bases suggesting that a putative proton transfer H-pathway pumps protons.
J.Biol.Chem., 299, 2023
|
|
7VUW
 
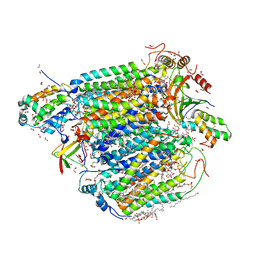 | | Bovine heart cytochrome c oxidase in the cyanide-bound fully oxidized state at 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Tsukihara, T. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic cyanide-probing for cytochrome c oxidase reveals structural bases suggesting that a putative proton transfer H-pathway pumps protons.
J.Biol.Chem., 299, 2023
|
|
9CT1
 
 | |
6MW2
 
 | |
7Z4O
 
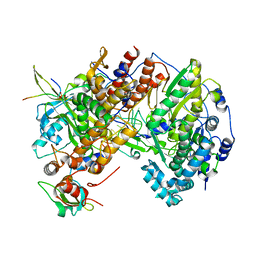 | |
9CR5
 
 | | Crystal structure of GNAT superfamily acetyltransferase PA2271 from Pseudomonas aeruginosa | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase PA2271, CHLORIDE ION, ... | | Authors: | Majorek, K.A, Cooper, D.R, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2024-07-20 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural, functional, and regulatory evaluation of a cysteine post-translationally modified Gcn5-related N-acetyltransferase.
Biochem.Biophys.Res.Commun., 748, 2025
|
|
7ZUN
 
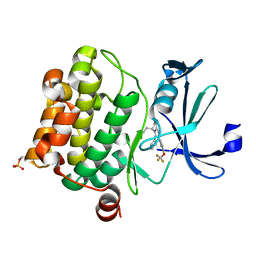 | | Crystal structure of PIM1 in complex with a Pyrrolo-Pyrazinone compound | | Descriptor: | (4~{S})-4-(2-azanylethyl)-6-phenyl-7-[3-(trifluoromethyloxy)phenyl]-3,4-dihydropyrrolo[1,2-a]pyrazin-1-ol, Isoform 2 of Serine/threonine-protein kinase pim-1 | | Authors: | Casale, E. | | Deposit date: | 2022-05-12 | | Release date: | 2022-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stereoselective synthesis of 3,4-dihydropyrrolo[1,2-a]pyrazin-1(2H)-one derivatives as PIM kinase inhibitors inspired from marine alkaloids.
Chirality, 34, 2022
|
|
9DM0
 
 | | Cryo-EM structure of the SFV009 3G01 Fab in complex with A/California/04/2009 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, ... | | Authors: | Fernandez Quintero, M.L, Ferguson, J.A, Han, J, Ward, A.B. | | Deposit date: | 2024-09-11 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structurally convergent antibodies derived from different vaccine strategies target the influenza virus HA anchor epitope with a subset of V H 3 and V K 3 genes.
Nat Commun, 16, 2025
|
|
2UUX
 
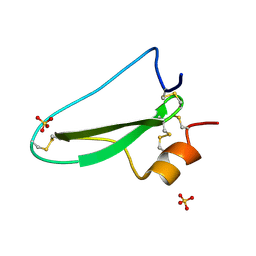 | | Structure of the tryptase inhibitor TdPI from a tick | | Descriptor: | SULFATE ION, TRYPTASE INHIBITOR | | Authors: | Siebold, C, Paesen, G.C, Harlos, K, Peacey, M.F, Nuttall, P.A, Stuart, D.I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Tick Protein with a Modified Kunitz Fold Inhibits Human Tryptase.
J.Mol.Biol., 368, 2007
|
|
5L8S
 
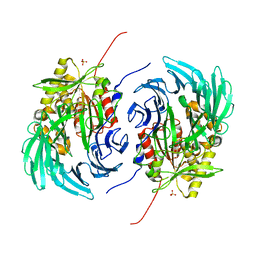 | | The crystal structure of a cold-adapted acylaminoacyl peptidase reveals a novel quaternary architecture based on the arm-exchange mechanism | | Descriptor: | Amino acyl peptidase, SULFATE ION | | Authors: | Brocca, S, Ferrari, C, Barbiroli, A, Pesce, A, Lotti, M, Nardini, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A bacterial acyl aminoacyl peptidase couples flexibility and stability as a result of cold adaptation.
FEBS J., 283, 2016
|
|
1A61
 
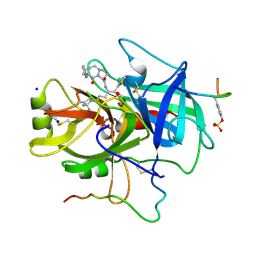 | | THROMBIN COMPLEXED WITH A BETA-MIMETIC THIAZOLE-CONTAINING INHIBITOR | | Descriptor: | (1S,7S)-7-amino-7-benzyl-N-{(1S)-4-carbamimidamido-1-[(S)-hydroxy(1,3-thiazol-2-yl)methyl]butyl}-8-oxohexahydro-1H-pyra zolo[1,2-a]pyridazine-1-carboxamide, ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), ... | | Authors: | St Charles, R, Matthews, J.H, Zhang, E, Tulinsky, A, Kahn, M. | | Deposit date: | 1998-03-05 | | Release date: | 1998-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bound structures of novel P3-P1' beta-strand mimetic inhibitors of thrombin.
J.Med.Chem., 42, 1999
|
|
9DFE
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with lasso peptide lariocidin and protein Y at 2.60A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Travin, D.Y, Jangra, M, Kaur, M, Darwish, L, Koteva, K, Klepacki, D, Wang, W, Tiffany, M, Sokaribo, A, Coombes, B.K, Vazquez-Laslop, N, Wright, G.D, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Broad Spectrum Lasso Peptide Antibiotic Targeting the Bacterial Ribosome.
Res Sq, 2024
|
|
8PN9
 
 | | Structure of human oligosaccharyltransferase OST-A complex bound to NGI-1 | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-[(1~{S},2~{R},3~{R},4~{R},5'~{S},6~{S},7~{R},8~{S},9~{R},12~{R},13~{R},15~{S},16~{S},18~{R})-5',7,9,13-tetramethyl-3,15-bis(oxidanyl)spiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icosane-6,2'-oxane]-16-yl]oxy-oxane-3,4,5-triol, (4R,7R)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(undecanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphadocosan-1-aminium, ... | | Authors: | Ramirez, A.S, Kowal, J, Locher, K.P. | | Deposit date: | 2023-06-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Positive selection CRISPR screens reveal a druggable pocket in an oligosaccharyltransferase required for inflammatory signaling to NF-kappa B.
Cell, 187, 2024
|
|
