3EF8
 
 | |
2ZBS
 
 | |
2ZJN
 
 | |
3VRC
 
 | | Crystal structure of cytochrome c' from Thermochromatium tepidum | | Descriptor: | CADMIUM ION, CHLORIDE ION, Cytochrome c', ... | | Authors: | Hirano, Y, Kimura, Y, Suzuki, H, Miki, K, Wang, Z.-Y. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure Analysis and Comparative Characterization of the Cytochrome c' and Flavocytochrome c from Thermophilic Purple Photosynthetic Bacterium Thermochromatium tepidum
Biochemistry, 51, 2012
|
|
3EIY
 
 | |
5Q0R
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N,1-dibenzyl-6-[(2-fluorophenyl)sulfonyl]-4,5,6,7-tetrahydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
2ZAS
 
 | | Crystal structure of human estrogen-related receptor gamma ligand binding domain complex with 4-alpha-cumylphenol, a bisphenol A derivative | | Descriptor: | 4-(1-methyl-1-phenylethyl)phenol, Estrogen-related receptor gamma, GLYCEROL | | Authors: | Matsushima, A, Kakuta, Y, Teramoto, T, Shimohigashi, Y. | | Deposit date: | 2007-10-09 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ERRgamma tethers strongly bisphenol A and 4-alpha-cumylphenol in an induced-fit manner
Biochem.Biophys.Res.Commun., 373, 2008
|
|
3VDX
 
 | |
3VEJ
 
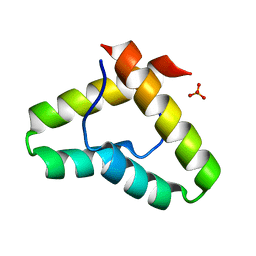 | | Crystal structure of the Get5 carboxyl domain from S. cerevisiae | | Descriptor: | PHOSPHATE ION, Ubiquitin-like protein MDY2 | | Authors: | Chartron, J.W, Vandervelde, D.G, Rao, M, Clemons Jr, W.M. | | Deposit date: | 2012-01-08 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Get5 Carboxyl-terminal Domain Is a Novel Dimerization Motif That Tethers an Extended Get4/Get5 Complex.
J.Biol.Chem., 287, 2012
|
|
5QCB
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, tert-butyl 4-(2-{3-[3-{[(3-methylbut-2-enoyl)amino]methyl}-4-(trifluoromethyl)phenyl]-1-[3-(morpholin-4-yl)propyl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}-2-oxoethyl)piperidine-1-carboxylate | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCS
 
 | | Crystal structure of BACE complex with BMC024 | | Descriptor: | (2R,4S)-N-BUTYL-4-HYDROXY-2-METHYL- 4-((E)-(4AS,12R,15S,17AS)-15-METHYL -14,17-DIOXO-2,3,4,4A,6,9,11,12,13, 14,15,16,17,17A-TETRADECAHYDRO-1H-5 ,10-DITHIA-1,13,16-TRIAZA-BENZOCYCL OPENTADECEN-12-YL)-BUTYRAMIDE, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QC3
 
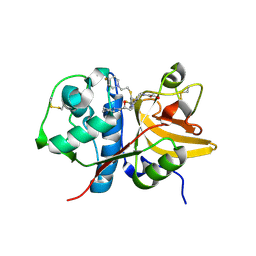 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 1-{3-[3-{[2-(4-fluoropiperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-1-[(2S)-2-hydroxy-3-(piperidin-1-yl)propyl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}-2-hydroxyethan-1-one, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCJ
 
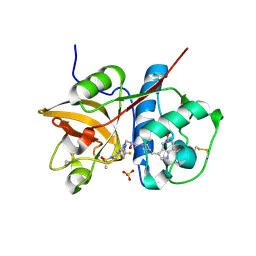 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 5-hydroxy-3-{1-[(2S)-2-hydroxy-3-{5-(methylsulfonyl)-3-[4-(trifluoromethyl)phenyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-1-yl}propyl]piperidin-4-yl}-1H-pyrrolo[3,2-c]pyridin-5-ium, Cathepsin S, SULFATE ION | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
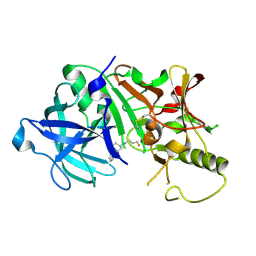
2ZJH
 
 | |
5QCC
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, tert-butyl 4-(2-{3-(4-chloro-3-iodophenyl)-1-[3-(morpholin-4-yl)propyl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}-2-oxoethyl)piperidine-1-carboxylate | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QJ4
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1827602749 | | Descriptor: | (2S,3S)-3-methyl-N-(1,2,3-thiadiazol-5-yl)tetrahydrofuran-2-carboxamide (non-preferred name), 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-11-12 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
2N2K
 
 | | Ensemble structure of the closed state of Lys63-linked diubiquitin in the absence of a ligand | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, ubiquitin | | Authors: | Liu, Z, Gong, Z, Tang, C. | | Deposit date: | 2015-05-10 | | Release date: | 2015-07-08 | | Last modified: | 2015-09-23 | | Method: | SOLUTION NMR | | Cite: | Lys63-linked ubiquitin chain adopts multiple conformational states for specific target recognition.
Elife, 4, 2015
|
|
3EI3
 
 | | Structure of the hsDDB1-drDDB2 complex | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, TETRAETHYLENE GLYCOL | | Authors: | Scrima, A, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
5QAU
 
 | | OXA-48 IN COMPLEX WITH COMPOUND 26a | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(1~{H}-1,2,3,4-tetrazol-5-yl)phenyl]benzoic acid, Beta-lactamase, ... | | Authors: | Lund, B.A, Leiros, H.K.S. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A focused fragment library targeting the antibiotic resistance enzyme - Oxacillinase-48: Synthesis, structural evaluation and inhibitor design.
Eur J Med Chem, 145, 2018
|
|
2ZYS
 
 | | A. Fulgidus lipase with fatty acid fragment and chloride | | Descriptor: | CHLORIDE ION, Lipase, putative, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
3EJN
 
 | |
5QC8
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, N-benzyl-1-{3-[(2-chloro-5-{5-(methylsulfonyl)-1-[3-(morpholin-4-yl)propyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]phenyl}methanamine | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
3EN8
 
 | |
3A26
 
 | | Crystal structure of P. horikoshii TYW2 in complex with MeSAdo | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, TETRAETHYLENE GLYCOL, Uncharacterized protein PH0793 | | Authors: | Umitsu, M, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2009-04-28 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of AdoMet-dependent aminocarboxypropyl transfer reaction catalyzed by tRNA-wybutosine synthesizing enzyme, TYW2
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
305D
 
 | |
