6JL9
 
 | | Crystal structure of a frog ependymin related protein | | Descriptor: | CALCIUM ION, Ependymin-related 1 | | Authors: | Park, S.Y. | | Deposit date: | 2019-03-04 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of three ependymin-related proteins suggest their function as a hydrophobic molecule binder.
Iucrj, 6, 2019
|
|
6JLE
 
 | | Crystal structure of MORN4/Myo3a complex | | Descriptor: | CITRIC ACID, GLYCEROL, MORN repeat-containing protein 4, ... | | Authors: | Li, J, Liu, H, Raval, M.H, Wan, J, Yengo, C.M, Liu, W, Zhang, M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the MORN4/Myo3a Tail Complex Reveals MORN Repeats as Protein Binding Modules.
Structure, 27, 2019
|
|
3CR3
 
 | | Structure of a transient complex between Dha-kinase subunits DhaM and DhaL from Lactococcus lactis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PTS-dependent dihydroxyacetone kinase, ... | | Authors: | Jeckelmann, J.M, Zurbriggen, A, Christen, S, Baumann, U, Erni, B. | | Deposit date: | 2008-04-04 | | Release date: | 2008-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structures of the Three Lactococcus lactis Dihydroxyacetone Kinase Subunits and of a Transient Intersubunit Complex.
J.Biol.Chem., 283, 2008
|
|
6JLI
 
 | | Crystal structure of CTLD7 domain of human PLA2R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Secretory phospholipase A2 receptor | | Authors: | Yu, B, Hu, Z, Kong, D, Cheng, C, He, Y. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.778 Å) | | Cite: | Crystal structure of the CTLD7 domain of human M-type phospholipase A2 receptor.
J.Struct.Biol., 207, 2019
|
|
6JGO
 
 | | Crystal structure of barley exohydrolaseI W434H mutant in complex with 4I,4III,4V-S-trithiocellohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JHA
 
 | | Crystal structure of NADPH bound AerF from Microcystis aeruginosa | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase family protein, TRIETHYLENE GLYCOL | | Authors: | Qiu, X. | | Deposit date: | 2019-02-17 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and functional investigation of AerF, a NADPH-dependent alkenal double bond reductase participating in the biosynthesis of Choi moiety of aeruginosin
J.Struct.Biol., 2019
|
|
6JHK
 
 | |
6JLO
 
 | | XFEL structure of cyanobacterial photosystem II (2F state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JHS
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F7 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JI5
 
 | | brd4-bd1 bound with ligand 167 | | Descriptor: | (3R)-4-cyclopentyl-6-[1-(2,4-dimethylphenyl)-3-(4-methylpiperazine-1-carbonyl)-1H-1,2,4-triazol-5-yl]-1,3-dimethyl-3,4-dihydroquinoxalin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Cao, D.Y, Li, Y.L, Du, Z.Y, Li, J, Xiong, B. | | Deposit date: | 2019-02-20 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | brd4-bd1 bound with ligand 167
To Be Published
|
|
6JID
 
 | | Human MTHFD2 in complex with Compound 1 | | Descriptor: | 5-(4-oxo-2-phenyl-1,5,7,8-tetrahydropyrido[4,3-d]pyrimidine-6(4H)-carbonyl)-1,3-dihydro-2H-2lambda~6~,1-benzothiazole-2,2-dione, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Suzuki, M, Matsui, Y, Matsuhashi, N, Kawai, J. | | Deposit date: | 2019-02-20 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design and Synthesis of an Isozyme-Selective MTHFD2 Inhibitor with a Tricyclic Coumarin Scaffold.
Acs Med.Chem.Lett., 10, 2019
|
|
6JIL
 
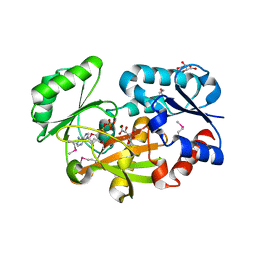 | | Crystal structure of D-cycloserine synthetase DcsG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cycloserine biosynthesis protein DcsG, L(+)-TARTARIC ACID, ... | | Authors: | Matoba, Y, Sugiyama, M. | | Deposit date: | 2019-02-22 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Cyclization mechanism catalyzed by an ATP-grasp enzyme essential for d-cycloserine biosynthesis.
Febs J., 287, 2020
|
|
6JO8
 
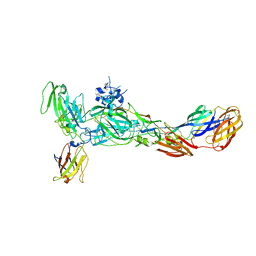 | | The complex structure of CHIKV envelope glycoprotein bound to human MXRA8 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHIKV E1, ... | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, F.G. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
6JPK
 
 | |
6JPV
 
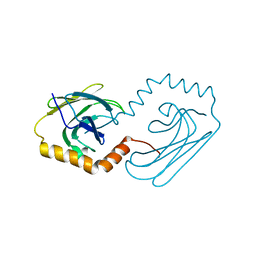 | | Structural analysis of AIMP2-DX2 and HSP70 interaction | | Descriptor: | Heat shock 70 kDa protein 1A,Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 | | Authors: | Cho, H.Y, Son, S.Y, Jeon, Y.H. | | Deposit date: | 2019-03-28 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15000653 Å) | | Cite: | Targeting the interaction of AIMP2-DX2 with HSP70 suppresses cancer development.
Nat.Chem.Biol., 16, 2020
|
|
6JIW
 
 | | Crystal structure of S. aureus CntK with C72S mutation | | Descriptor: | 1,2-ETHANEDIOL, Diaminopimelate epimerase, SULFATE ION | | Authors: | Luo, S, Ju, Y, Zhou, H. | | Deposit date: | 2019-02-23 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of CntK, the cofactor-independent histidine racemase in staphylopine-mediated metal acquisition of Staphylococcus aureus.
Int.J.Biol.Macromol., 135, 2019
|
|
3CSJ
 
 | | Human glutathione s-transferase p1-1 in complex with chlorambucil | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORAMBUCIL, CHLORIDE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The anti-cancer drug chlorambucil as a substrate for the human polymorphic enzyme glutathione transferase P1-1: kinetic properties and crystallographic characterisation of allelic variants.
J.Mol.Biol., 380, 2008
|
|
6JJ0
 
 | |
6JJB
 
 | | BRD4 in complex with ZZM1 | | Descriptor: | 2-methoxy-N-(1-methyl-2-oxidanylidene-benzo[cd]indol-6-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Xu, J, Chen, Y, Jiang, F, Zhu, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.508 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones and Pyrrolo[4,3,2-de]quinolin-2(1H)-ones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with Selectivity for the First Bromodomain with Potential High Efficiency against Acute Gouty Arthritis.
J.Med.Chem., 62, 2019
|
|
6JJE
 
 | |
6JQ8
 
 | |
6JQL
 
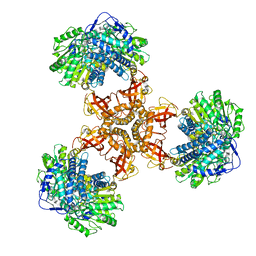 | | Structure of PaaZ, a bifunctional enzyme | | Descriptor: | Bifunctional protein PaaZ | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JRQ
 
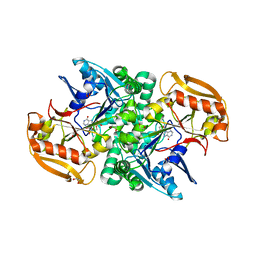 | | Crystal structure of adenylosuccinate synthetase, PurA, from Thermus thermophilus | | Descriptor: | 1,2-ETHANEDIOL, Adenylosuccinate synthetase, INOSINIC ACID | | Authors: | Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2019-04-05 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of adenylosuccinate synthetase, PurA, from Thermus thermophilus HB8
To Be Published
|
|
6JJF
 
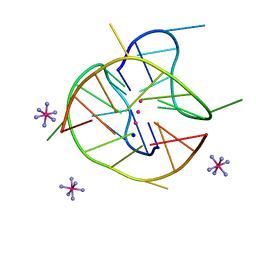 | | Crystal structure of a two-quartet DNA mixed-parallel/antiparallel G-quadruplex | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*GP*CP*TP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3'), POTASSIUM ION, ... | | Authors: | Zhang, Y.S, EI Omari, K, Duman, R, Wagner, A, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
6JJP
 
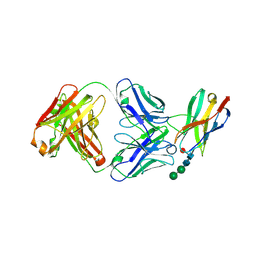 | | Crystal structure of Fab of a PD-1 monoclonal antibody MW11-h317 in complex with PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of MW11-h317, Programmed cell death protein 1, ... | | Authors: | Wang, M, Wang, J, Wang, R, Jiao, S, Wang, S, Zhang, J, Zhang, M. | | Deposit date: | 2019-02-26 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of a monoclonal antibody that targets PD-1 in a manner requiring PD-1 Asn58 glycosylation.
Commun Biol, 2, 2019
|
|
