3NOJ
 
 | | The structure of HMG/CHA aldolase from the protocatechuate degradation pathway of Pseudomonas putida | | Descriptor: | 4-carboxy-4-hydroxy-2-oxoadipate aldolase/oxaloacetate decarboxylase, MAGNESIUM ION, PYRUVIC ACID, ... | | Authors: | Kimber, M.S, Wang, W, Mazurkewich, S, Seah, S.Y.K. | | Deposit date: | 2010-06-25 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and Kinetic Characterization of 4-Hydroxy-4-methyl-2-oxoglutarate/4-Carboxy-4-hydroxy-2-oxoadipate Aldolase, a Protocatechuate Degradation Enzyme Evolutionarily Convergent with the HpaI and DmpG Pyruvate Aldolases.
J.Biol.Chem., 285, 2010
|
|
3NQO
 
 | |
4Q9C
 
 | | IgNAR antibody domain C3 | | Descriptor: | CHLORIDE ION, Novel antigen receptor, SODIUM ION, ... | | Authors: | Feige, J.M, Graewert, M.A, Marcinowski, M, Hennig, J, Behnke, J, Auslaender, D, Herold, E.M, Peschek, J, Castro, C.D, Flajnik, M.F, Hendershot, L.M, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3SWS
 
 | |
4QJ3
 
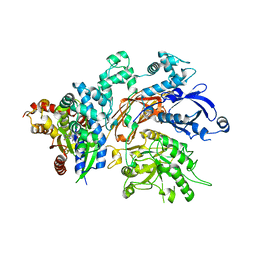 | | Structure of a fragment of human phospholipase C-beta3 delta472-559, in complex with Galphaq | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lyon, A.M, Tesmer, J.J.G. | | Deposit date: | 2014-06-03 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular mechanisms of phospholipase C beta 3 autoinhibition.
Structure, 22, 2014
|
|
2A5I
 
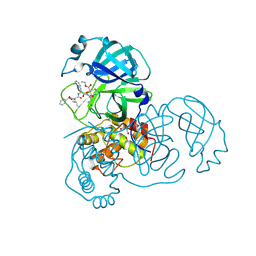 | | Crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide in the space group C2 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 1,2-ETHANEDIOL, 3C-like peptidase, ... | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
1GVV
 
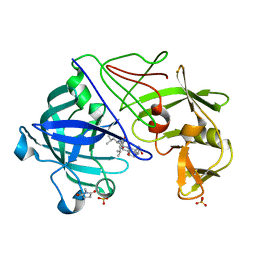 | | Five Atomic Resolution Structures of Endothiapepsin Inhibitor Complexes; implications for the Aspartic Proteinase Mechanism | | Descriptor: | ENDOTHIAPEPSIN, N-[(5S,9S,10S,13S)-9-hydroxy-5,10-bis(2-methylpropyl)-4,7,12,16-tetraoxo-3,6,11,17-tetraazabicyclo[17.3.1]tricosa-1(23),19,21-trien-13-yl]-3-(naphthalen-1-yl)-2-(naphthalen-1-ylmethyl)propanamide, SULFATE ION | | Authors: | Coates, L, Erskine, P.T, Crump, M.P, Wood, S.P, Cooper, J.B. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Five Atomic Resolution Structures of Endothiapepsin Inhibitor Complexes: Implications for the Aspartic Proteinase Mechanism
J.Mol.Biol., 318, 2002
|
|
4HOS
 
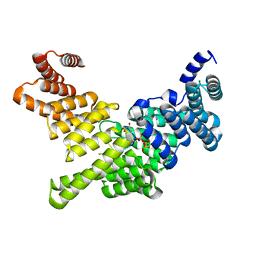 | | Crystal Structure of Full-Length Human IFIT5 with 5`-triphosphate Oligouridine | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 5, RNA (5'-R(*(UTP)P*UP*UP*U)-3'), SODIUM ION | | Authors: | Abbas, Y.M, Pichlmair, A, Gorna, M.W, Superti-Furga, G, Nagar, B. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for viral 5'-PPP-RNA recognition by human IFIT proteins.
Nature, 494, 2013
|
|
1U0M
 
 | | Crystal Structure of 1,3,6,8-Tetrahydroxynaphthalene Synthase (THNS) from Streptomyces coelicolor A3(2): a Bacterial Type III Polyketide Synthase (PKS) Provides Insights into Enzymatic Control of Reactive Polyketide Intermediates | | Descriptor: | GLYCEROL, POLYETHYLENE GLYCOL (N=34), putative polyketide synthase | | Authors: | Austin, M.B, Izumikawa, M, Bowman, M.E, Udwary, D.W, Ferrer, J.L, Moore, B.S, Noel, J.P. | | Deposit date: | 2004-07-13 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of a bacterial type III polyketide synthase and enzymatic control of reactive polyketide intermediates
J.Biol.Chem., 279, 2004
|
|
4HVK
 
 | | Crystal structure and functional studies of an unusual L-cysteine desulfurase from Archaeoglobus fulgidus. | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yamanaka, Y, Zeppieri, L, Nicolet, Y, Marinoni, E.N, de Oliveira, J.S, Masafumi, O, Dean, D.R, Fontecilla-Camps, J.C. | | Deposit date: | 2012-11-06 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure and functional studies of an unusual L-cysteine desulfurase from Archaeoglobus fulgidus.
Dalton Trans, 42, 2013
|
|
5O08
 
 | | Crystal structure of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus in complex with Dephospho-coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, DEPHOSPHO COENZYME A, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
5O0D
 
 | | Crystal structure of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus in complex with 3-Phenoxymandelic acid (Fragment 4) | | Descriptor: | (2~{S})-2-oxidanyl-2-(3-phenoxyphenyl)ethanoic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
4I5U
 
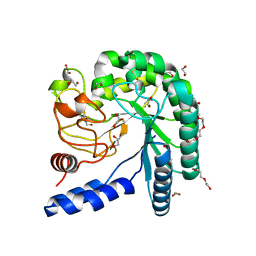 | | Crystal structure of a fungal chimeric cellobiohydrolase Cel6A | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Chimeric cel6A, ... | | Authors: | Arnold, F.H, Wu, I. | | Deposit date: | 2012-11-29 | | Release date: | 2013-04-03 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Engineered thermostable fungal Cel6A and Cel7A cellobiohydrolases hydrolyze cellulose efficiently at elevated temperatures.
Biotechnol.Bioeng., 110, 2013
|
|
2A5K
 
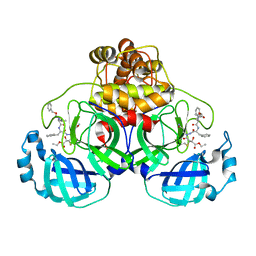 | | Crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide in space group P212121 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 3C-like peptidase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
3NWP
 
 | |
4E84
 
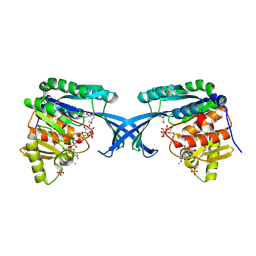 | | Crystal Structure of Burkholderia cenocepacia HldA | | Descriptor: | 1,7-di-O-phosphono-D-glycero-beta-D-manno-heptopyranose, 7-O-phosphono-D-glycero-beta-D-manno-heptopyranose, CHLORIDE ION, ... | | Authors: | Lee, T.-W, Junop, M.S. | | Deposit date: | 2012-03-19 | | Release date: | 2012-12-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural-functional studies of Burkholderia cenocepacia D-glycero-beta-D-manno-heptose 7-phosphate kinase (HldA) and characterization of inhibitors with antibiotic adjuvant and antivirulence properties.
J.Med.Chem., 56, 2013
|
|
4QXX
 
 | |
4D9J
 
 | |
3NXQ
 
 | | Angiotensin Converting Enzyme N domain glycsoylation mutant (Ndom389) in complex with RXP407 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, CHLORIDE ION, ... | | Authors: | Anthony, C.S, Corradi, H.R, Schwager, S.L.U, Redelinghuys, P, Georgiadis, D, Dive, V, Acharya, K.R, Sturrock, E.D. | | Deposit date: | 2010-07-14 | | Release date: | 2010-09-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The N domain of human angiotensin-I-converting enzyme: the role of N-glycosylation and the crystal structure in complex with an N domain-specific phosphinic inhibitor, RXP407.
J.Biol.Chem., 285, 2010
|
|
3SR0
 
 | |
3T34
 
 | | Arabidopsis thaliana dynamin-related protein 1A (AtDRP1A) in prefission state | | Descriptor: | Dynamin-related protein 1A, LINKER, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yan, L.M, Ma, Y.Y, Sun, Y.N, Lou, Z.Y. | | Deposit date: | 2011-07-24 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structural basis for mechanochemical role of Arabidopsis thaliana dynamin-related protein in membrane fission
J Mol Cell Biol, 3, 2011
|
|
3FDH
 
 | |
3FYZ
 
 | | OXA-24 beta-lactamase complex with SA4-17 inhibitor | | Descriptor: | (2S,3R)-2-[(7-aminocarbonyl-2-methanoyl-indolizin-3-yl)amino]-4-aminocarbonyloxy-3-methyl-3-sulfino-butanoic acid, Beta-lactamase OXA-24, TETRAETHYLENE GLYCOL | | Authors: | Romero, A, Santillana, E. | | Deposit date: | 2009-01-23 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase.
J.Am.Chem.Soc., 132, 2010
|
|
3TAS
 
 | | Small laccase from Streptomyces viridosporus T7A | | Descriptor: | ACETATE ION, COPPER (II) ION, OXYGEN MOLECULE, ... | | Authors: | Lukk, T, Majumdar, S, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2011-08-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Roles of small laccases from Streptomyces in lignin degradation.
Biochemistry, 53, 2014
|
|
4DFK
 
 | | large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with 5-(N-(10-hydroxydecanoyl)-aminopentinyl)-2-dUTP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5-{5-[(10-hydroxydecanoyl)amino]pent-1-yn-1-yl}uridine 5'-(tetrahydrogen triphosphate), 5'-d(AAAAGGCGCCGTGGTC)-3', ... | | Authors: | Bergen, K, Steck, A, Struett, S, Baccaro, A, Welte, W, Diederichs, K, Marx, A. | | Deposit date: | 2012-01-24 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Structures of KlenTaq DNA Polymerase Caught While Incorporating C5-Modified Pyrimidine and C7-Modified 7-Deazapurine Nucleoside Triphosphates.
J.Am.Chem.Soc., 134, 2012
|
|
