3ARZ
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with 2-(imidazolin-2-yl)-5-isothiocyanatobenzofuran | | Descriptor: | 2-(5-isothiocyanato-1-benzofuran-2-yl)-4,5-dihydro-1H-imidazole, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
4I5H
 
 | |
4IB2
 
 | |
3AVL
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3ARV
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with Sanguinarine | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ASD
 
 | | MamA R50E mutant | | Descriptor: | MamA | | Authors: | Zeytuni, N, Davidov, G, Zarivach, R. | | Deposit date: | 2010-12-10 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Self-recognition mechanism of MamA, a magnetosome-associated TPR-containing protein, promotes complex assembly
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AV4
 
 | | Crystal structure of mouse DNA methyltransferase 1 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AW9
 
 | |
3ARR
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with PENTOXIFYLLINE | | Descriptor: | 3,7-DIMETHYL-1-(5-OXOHEXYL)-3,7-DIHYDRO-1H-PURINE-2,6-DIONE, Chitinase A | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3AS2
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with Propentofylline | | Descriptor: | 3-methyl-1-(5-oxohexyl)-7-propyl-3,7-dihydro-1H-purine-2,6-dione, Chitinase A | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ASS
 
 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with Lewis-b | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
4IAJ
 
 | |
3ATT
 
 | | Crystal structure of Rv3168 with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Kim, Y.-G, Kim, S, Nguyen, C.M.T, Kim, K.-J. | | Deposit date: | 2011-01-13 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv3168: a putative aminoglycoside antibiotics resistance enzyme
Proteins, 79, 2011
|
|
3AV9
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-02 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVF
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
4HSP
 
 | |
3ACF
 
 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in a ligand-free form | | Descriptor: | Beta-1,4-endoglucanase, CALCIUM ION, SULFATE ION | | Authors: | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
3ACP
 
 | | Crystal Structure of Yeast Rpn14, a Chaperone of the 19S Regulatory Particle of the Proteasome | | Descriptor: | WD repeat-containing protein YGL004C | | Authors: | Kim, S, Saeki, Y, Suzuki, A, Takagi, K, Fukunaga, K, Yamane, T, Kato, K, Tanaka, K, Mizushima, T. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of yeast Rpn14, a chaperone of the 19S regulatory particle of the proteasome
J.Biol.Chem., 285, 2010
|
|
3AE3
 
 | | Crystal structure of porcine heart mitochondrial complex II bound with 2-Nitro-N-phenyl-benzamide | | Descriptor: | 2-nitro-N-phenylbenzamide, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Sasaki, T, Shindo, M, Kido, Y, Inaoka, D.K, Omori, J, Osanai, A, Sakamoto, K, Mao, J, Matsuoka, S, Inoue, M, Honma, T, Tanaka, A, Kita, K. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structure of porcine heart mitochondrial complex II bound with 2-Nitro-N-phenyl-benzamide
To be Published
|
|
3AEM
 
 | | Reaction intermediate structure of Entamoeba histolytica methionine gamma-lyase 1 containing Michaelis complex and methionine imine-pyridoxamine-5'-phosphate | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-4-(methylsulfanyl)butanoic acid, GLYCEROL, METHIONINE, ... | | Authors: | Karaki, T, Sato, D, Shimizu, A, Nozaki, T, Harada, S. | | Deposit date: | 2010-02-10 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Entamoeba histolytica methionine gamma-lyase 1
To be published
|
|
3AEE
 
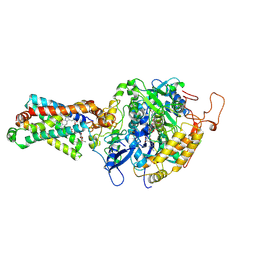 | | Crystal structure of porcine heart mitochondrial complex II bound with Atpenin A5 | | Descriptor: | 3-[(2S,4S,5R)-5,6-DICHLORO-2,4-DIMETHYL-1-OXOHEXYL]-4-HYDROXY-5,6-DIMETHOXY-2(1H)-PYRIDINONE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Sasaki, T, Shindo, M, Kido, Y, Inaoka, D.K, Omori, J, Osanai, A, Sakamoto, K, Mao, J, Matsuoka, S, Inoue, M, Honma, T, Tanaka, A, Kita, K. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Crystal structure of porcine heart mitochondrial complex II bound with Atpenin A5
To be Published
|
|
4HUM
 
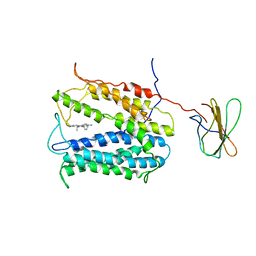 | | MATE transporter NorM-NG in complex with ethidium and monobody | | Descriptor: | ETHIDIUM, Multidrug efflux protein, Protein B | | Authors: | Lu, M. | | Deposit date: | 2012-11-02 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structures of a Na+-coupled, substrate-bound MATE multidrug transporter.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4I2A
 
 | |
4I2G
 
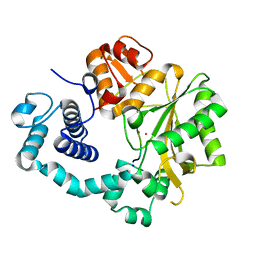 | | Binary complex of mouse TdT with ssDNA | | Descriptor: | 5'-D(P*AP*AP*AP*AP*A)-3', COBALT (II) ION, DNA nucleotidylexotransferase, ... | | Authors: | Gouge, J, Delarue, M. | | Deposit date: | 2012-11-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Intermediates along the Catalytic Cycle of Terminal Deoxynucleotidyltransferase: Dynamical Aspects of the Two-Metal Ion Mechanism.
J.Mol.Biol., 425, 2013
|
|
375D
 
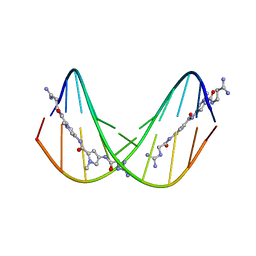 | | A NOVEL END-TO-END BINDING OF TWO NETROPSINS TO THE DNA DECAMER D(CCCCCIIIII)2 | | Descriptor: | DNA (5'-D(*CP*CP*CP*CP*CP*IP*IP*IP*IP*I)-3'), NETROPSIN | | Authors: | Chen, X, Rao, S.T, Sekar, K, Sundaralingam, M. | | Deposit date: | 1998-01-14 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Novel End-to-End Binding of Two Netropsins to the DNA Decamers d(CCCCCIIIII) 2, d(CCCBr5CCIIIII)2, d(CBr5CCCCIIIII)2
Nucleic Acids Res., 26, 1998
|
|
