4URD
 
 | | Cryo-EM map of Trigger Factor bound to a translating ribosome | | Descriptor: | TRIGGER FACTOR | | Authors: | Deeng, J, Chan, K.Y, van der Sluis, E, Bischoff, L, Berninghausen, O, Han, W, Gumbart, J, Schulten, K, Beatrix, B, Beckmann, R. | | Deposit date: | 2014-06-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Dynamic Behavior of Trigger Factor on the Ribosome.
J.Mol.Biol., 428, 2016
|
|
6M1S
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12o | | Descriptor: | 3-[5-[8-(ethylamino)-6-fluoranyl-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]oxy-2,2-dimethyl-propanoic acid, CHLORIDE ION, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
6LRB
 
 | |
6M35
 
 | | Crystal structure of sulfur oxygenase reductase from Sulfurisphaera tokodaii | | Descriptor: | FE (III) ION, GLYCEROL, SULFATE ION, ... | | Authors: | Sato, Y, Yabuki, T, Arakawa, T, Yamada, C, Fushinobu, S, Wakagi, T. | | Deposit date: | 2020-03-02 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystallographic and cryogenic electron microscopic structures and enzymatic characterization of sulfur oxygenase reductase fromSulfurisphaera tokodaii.
J Struct Biol X, 4, 2020
|
|
6LRY
 
 | | Crystal structure of human endothelin ETB receptor in complex with sarafotoxin S6b | | Descriptor: | Endothelin receptor type B,Endolysin,Endothelin receptor type B, Sarafotoxin-B | | Authors: | Izume, T, Miyauchi, H, Shihoya, W, Nureki, O. | | Deposit date: | 2020-01-16 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human endothelin ETBreceptor in complex with sarafotoxin S6b.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
6LST
 
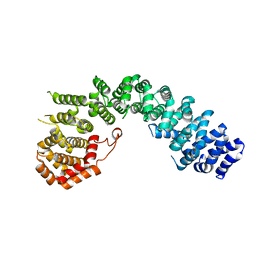 | | Crystal straucture of Uso1-1 | | Descriptor: | Intracellular protein transport protein USO1 | | Authors: | Heo, Y.Y, Lee, H.H. | | Deposit date: | 2020-01-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structures of Uso1 membrane tether reveal an alternative conformation in the globular head domain
Sci Rep, 10, 2020
|
|
4V1W
 
 | |
6MB0
 
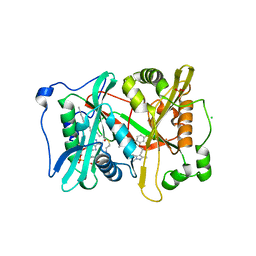 | |
6MBJ
 
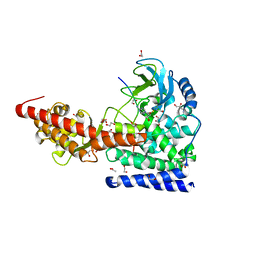 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, P21 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin Peptide, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
6LV0
 
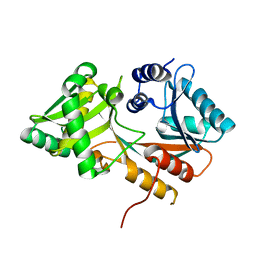 | |
6LQ4
 
 | |
4WCT
 
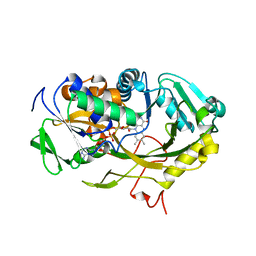 | | The crystal structure of Fructosyl amine: oxygen oxidoreductase (Amadoriase I) from Aspergillus fumigatus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine:oxygen oxidoreductase | | Authors: | Rigoldi, F, Gautieri, A, Dalle Vedove, A, Lucarelli, A.P, Vesentini, S, Parisini, E. | | Deposit date: | 2014-09-05 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the deglycating enzyme Amadoriase I in its free form and substrate-bound complex.
Proteins, 84, 2016
|
|
4WD9
 
 | |
6M1U
 
 | |
6M3R
 
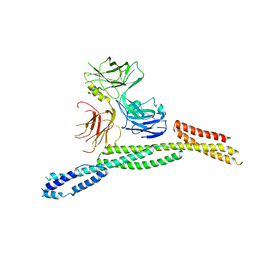 | | Crystal structure of AnkG/beta4-spectrin complex | | Descriptor: | Ankyrin-3, Spectrin beta chain | | Authors: | Li, J, Chen, K, Zhu, R, Zhang, M. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.313 Å) | | Cite: | Structural Basis Underlying Strong Interactions between Ankyrins and Spectrins.
J.Mol.Biol., 432, 2020
|
|
6M5A
 
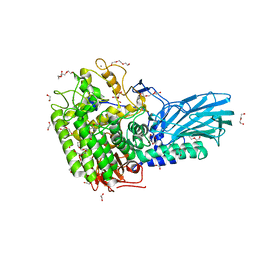 | | Crystal structure of GH121 beta-L-arabinobiosidase HypBA2 from Bifidobacterium longum | | Descriptor: | 1,2-ETHANEDIOL, Beta-L-arabinobiosidase, CALCIUM ION, ... | | Authors: | Saito, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2020-03-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of beta-L-arabinobiosidase belonging to glycoside hydrolase family 121.
Plos One, 15, 2020
|
|
6M85
 
 | |
6LLF
 
 | | Biphenyl-2,2',3-triol-soaked resting complex of Oxy and Fd in carbazole 1,9a-dioxygenase | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-hydroxyphenyl)benzene-1,2-diol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, Y.X, Suzuki-Minakuchi, C, Nojiri, H. | | Deposit date: | 2019-12-23 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Biphenyl-2,2',3-triol-soaked resting complex of Oxy and Fd in carbazole 1,9a-dioxygenase
To Be Published
|
|
6M9K
 
 | |
6MAI
 
 | |
6MB1
 
 | | Crystal structure of N-myristoyl transferase (NMT) from Plasmodium vivax in complex with inhibitor IMP-1002 | | Descriptor: | 1,2-ETHANEDIOL, 1-(5-{4-fluoro-2-[2-(1,3,5-trimethyl-1H-pyrazol-4-yl)ethoxy]phenyl}-1-methyl-1H-indazol-3-yl)-N,N-dimethylmethanamine, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-08-29 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Identification of Resistance Breaking Antimalarial N‐Myristoyltransferase Inhibitors.
Cell Chem Biol, 26, 2019
|
|
4UYJ
 
 | | Crystal structure of a Signal Recognition Particle Alu domain in the elongation arrest conformation | | Descriptor: | SIGNAL RECOGNITION PARTICLE 14 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, SRP RNA | | Authors: | Bousset, L, Mary, C, Brooks, M.A, Scherrer, A, Strub, K, Cusack, S. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structure of a Signal Recognition Particle Alu Domain in the Elongation Arrest Conformation.
RNA, 20, 2014
|
|
6MBH
 
 | |
6LP5
 
 | | Structure of Sinonovacula constricta ferritin | | Descriptor: | FE (II) ION, FE (III) ION, Ferritin, ... | | Authors: | Su, X.R, Ming, T.H, Su, C. | | Deposit date: | 2020-01-08 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystallographic characterization of ferritin from Sinonovacula constricta.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
6MBW
 
 | | Structure of Transcription Factor | | Descriptor: | Signal transducer and activator of transcription 5B | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2018-08-30 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural and functional consequences of the STAT5BN642H driver mutation.
Nat Commun, 10, 2019
|
|
