4E5G
 
 | | Crystal structure of avian influenza virus PAn bound to compound 2 | | Descriptor: | 2-4-DIOXO-4-PHENYLBUTANOIC ACID, MANGANESE (II) ION, Polymerase protein PA, ... | | Authors: | DuBois, R.M, Slavish, P.J, Webb, T.R, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.647 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
4FO5
 
 | |
2YG2
 
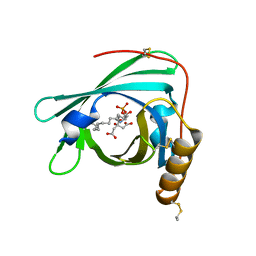 | | Structure of apolioprotein M in complex with Sphingosine 1-Phosphate | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, APOLIPOPROTEIN M, CITRATE ANION | | Authors: | Christoffersen, C, Obinata, H, Gowda, S, Galvani, S, Ahnstrom, J, Sevvana, M, Egerer-Sieber, C, Muller, Y.A, Hla, T, Nielsen, L, Dahlback, B. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Endothelium-Protective Sphingosine-1-Phosphate Provided by Hdl-Associated Apolipoprotein M.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4FE5
 
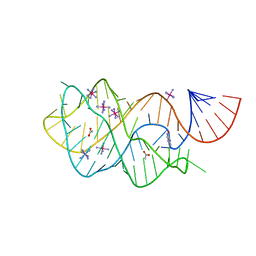 | | Crystal structure of the xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | | Descriptor: | ACETATE ION, COBALT HEXAMMINE(III), HYPOXANTHINE, ... | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-29 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine.
Nature, 432, 2004
|
|
4FEO
 
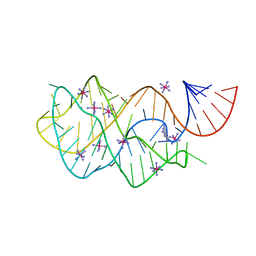 | | Crystal structure of the AU25A/A46G/C74U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with 2,6-diaminopurine | | Descriptor: | 9H-PURINE-2,6-DIAMINE, COBALT HEXAMMINE(III), U25A/A46G/C74U mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
4FDW
 
 | |
4FMR
 
 | |
4FHO
 
 | |
4H3P
 
 | | Crystal structure of human ERK2 complexed with a MAPK docking peptide | | Descriptor: | Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6 kinase alpha-1 | | Authors: | Gogl, G, Toeroe, I, Remenyi, A. | | Deposit date: | 2012-09-14 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein-peptide complex crystallization: a case study on the ERK2 mitogen-activated protein kinase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GPV
 
 | |
2XH6
 
 | | Clostridium perfringens enterotoxin | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, HEAT-LABILE ENTEROTOXIN B CHAIN, octyl beta-D-glucopyranoside | | Authors: | Briggs, D.C, Naylor, C.E, Smedley III, J.G, MCClane, B.A, Basak, A.K. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of the Food-Poisoning Clostridium Perfringens Enterotoxin Reveals Similarity to the Aerolysin-Like Pore-Forming Toxins
J.Mol.Biol., 413, 2011
|
|
4HAG
 
 | | Crystal structure of fc-fragment of human IgG2 antibody (centered crystal form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-2 chain C region | | Authors: | Teplyakov, A, Malia, T, Obmolova, G, Zhao, Y, Gilliland, G. | | Deposit date: | 2012-09-26 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | IgG2 Fc structure and the dynamic features of the IgG CH2-CH3 interface.
Mol.Immunol., 56, 2013
|
|
2XEI
 
 | | Human glutamate carboxypeptidase II in complex with Antibody- Recruiting Molecule ARM-P2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhang, A.X, Murelli, R.P, Barinka, C, Michel, J, Cocleaza, A, Jorgensen, W.L, Lubkowski, J, Spiegel, D.A. | | Deposit date: | 2010-05-14 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A Remote Arene-Binding Site on Prostate Specific Membrane Antigen Revealed by Antibody-Recruiting Small Molecules.
J.Am.Chem.Soc., 132, 2010
|
|
4GXY
 
 | | RNA structure | | Descriptor: | Adenosylcobalamin, Adenosylcobalamin riboswitch, IRIDIUM HEXAMMINE ION, ... | | Authors: | Serganov, A, Peselis, A. | | Deposit date: | 2012-09-04 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into ligand binding and gene expression control by an adenosylcobalamin riboswitch.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2ZY0
 
 | | Crystal structure of the human RXR alpha ligand binding domain bound to a synthetic agonist compound and a coactivator peptide | | Descriptor: | 4-[2-(1,1,3,3-tetramethyl-2,3-dihydro-1H-1,3-benzodisilol-5-yl)-1,3-dioxolan-2-yl]benzoic acid, GRIP1 from Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Sato, Y, Antony, P, Rochel, N, Moras, D, Structural Genomics Consortium for Research on Gene Expression (SGCGES) | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Silicon analogues of the RXR-selective retinoid agonist SR11237 (BMS649): chemistry and biology
Chemmedchem, 4, 2009
|
|
3AEG
 
 | | Crystal structure of porcine heart mitochondrial complex II bound with N-Biphenyl-3-yl-2-iodo-benzamide | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Harada, S, Sasaki, T, Shindo, M, Kido, Y, Inaoka, D.K, Omori, J, Osanai, A, Sakamoto, K, Mao, J, Matsuoka, S, Inoue, M, Honma, T, Tanaka, A, Kita, K. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Crystal structure of porcine heart mitochondrial complex II bound with N-Biphenyl-3-yl-2-iodo-benzamide
To be Published
|
|
4GOK
 
 | | The Crystal structure of Arl2GppNHp in complex with UNC119a | | Descriptor: | ADP-ribosylation factor-like protein 2, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Ismail, S, Xiang-Chen, Y, Miertzschke, M, Vetter, I, Koerner, C, Wittinghofer, A. | | Deposit date: | 2012-08-20 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for Arl3-specific release of myristoylated ciliary cargo from UNC119.
Embo J., 31, 2012
|
|
4GOQ
 
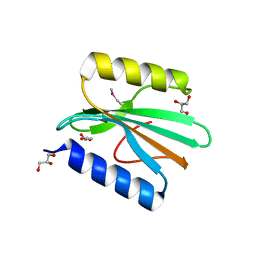 | |
358D
 
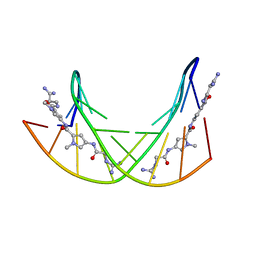 | |
3A0S
 
 | | PAS domain of histidine kinase ThkA (TM1359) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Sensor protein, TETRAETHYLENE GLYCOL, ... | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3A0Y
 
 | | Catalytic domain of histidine kinase ThkA (TM1359) (nucleotide free form 3: 1,2-propanediol, orthorombic) | | Descriptor: | Sensor protein | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-25 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3A0W
 
 | | Catalytic domain of histidine kinase ThkA (TM1359) for MAD phasing (nucleotide free form 2, orthorombic) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(ETHYLMERCURI-THIO)-BENZOIC ACID, ETHYL MERCURY ION, ... | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3A17
 
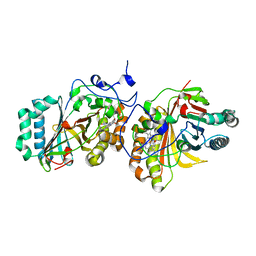 | | Crystal Structure of Aldoxime Dehydratase (OxdRE) in Complex with Butyraldoxime (Co-crystal) | | Descriptor: | (1Z)-butanal oxime, Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sawai, H, Sugimoto, H, Kato, Y, Asano, Y, Shiro, Y, Aono, S. | | Deposit date: | 2009-03-26 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of michaelis complex of aldoxime dehydratase
J.Biol.Chem., 284, 2009
|
|
3A4C
 
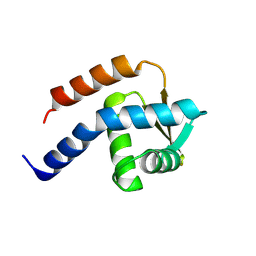 | | Crystal structure of cdt1 C terminal domain | | Descriptor: | DNA replication factor Cdt1 | | Authors: | Cho, Y, Lee, J.H. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Structure of the Cdt1 C-terminal domain: Conservation of the winged helix fold in replication licensing factors
Protein Sci., 18, 2009
|
|
3A4I
 
 | | Crystal structure of GMP synthetase PH1347 from Pyrococcus horikoshii OT3 | | Descriptor: | GMP synthase [glutamine-hydrolyzing] subunit B | | Authors: | Maruoka, S, Horita, S, Lee, W.C, Nagata, K, Tanokura, M. | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the ATPPase subunit and its substrate-dependent association with the GATase Subunit: a novel regulatory mechanism for a two-subunit-type GMP synthetase from Pyrococcus horikoshii OT3.
J.Mol.Biol., 395, 2010
|
|
