5AL0
 
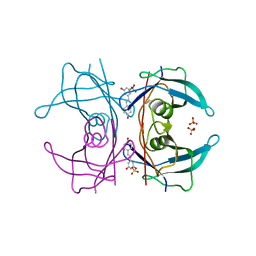 | | Transthyretin binding heterogeneity and anti-amyloidogenic activity of natural polyphenols and their metabolites: resveratrol-3-O-sulfate | | Descriptor: | DIMETHYL SULFOXIDE, RESVERATROL-3-O-SULFATE, SULFATE ION, ... | | Authors: | Florio, P, Foll, C, Cianci, M, Del Rio, D, Zanotti, G, Berni, R. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.388 Å) | | Cite: | Transthyretin Binding Heterogeneity and Anti-Amyloidogenic Activity of Natural Polyphenols and Their Metabolites
J.Biol.Chem., 290, 2015
|
|
8ITG
 
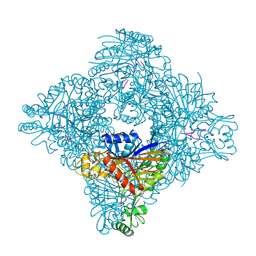 | |
5A71
 
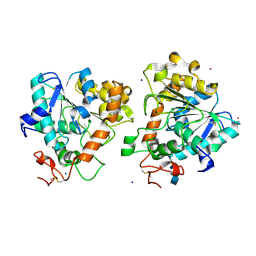 | | Open and closed conformations and protonation states of Candida antarctica Lipase B: atomic resolution native | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ISOPROPYL ALCOHOL, LIPASE B, ... | | Authors: | Stauch, B, Fisher, S.J, Cianci, M. | | Deposit date: | 2015-07-02 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Open and Closed States of Candida Antarctica Lipase B: Protonation and the Mechanism of Interfacial Activation.
J.Lipid Res., 56, 2015
|
|
8J53
 
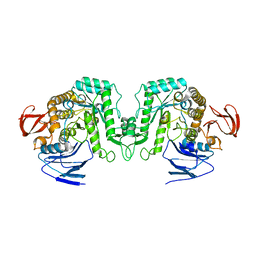 | |
8J51
 
 | |
8IQB
 
 | |
5ACB
 
 | | Crystal Structure of the Human Cdk12-Cyclink Complex | | Descriptor: | CYCLIN-DEPENDENT KINASE 12, CYCLIN-K, N-[4-[(3R)-3-[[5-chloranyl-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]piperidin-1-yl]carbonylphenyl]-4-(dimethylamino)butanamide | | Authors: | Dixon Clarke, S.E, Elkins, J.M, Pike, A.C.W, Mackenzie, A, Goubin, S, Strain-Damerell, C, Mahajan, P, Tallant, C, Chalk, R, Wiggers, H, Kopec, J, Fitzpatrick, F, Burgess-Brown, N, Carpenter, E.P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-08-14 | | Release date: | 2016-06-15 | | Last modified: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Covalent Targeting of Remote Cysteine Residues to Develop Cdk12 and Cdk13 Inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
8IQD
 
 | |
8J50
 
 | |
8J52
 
 | | Crystal structure of Flavihumibacter petaseus GH31 alpha-galactosidase mutant D304A in complex with alpha-1,4-galactobiose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GH31 alpha-galactosidase, ... | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2023-04-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function analysis of bacterial GH31 alpha-galactosidases specific for alpha-(1→4)-galactobiose.
Febs J., 290, 2023
|
|
5AB5
 
 | |
5ADG
 
 | | Structure of human nNOS R354A G357D mutant heme domain in complex with 7-((4-Chloro-3-((methylamino)methyl)phenoxy)methyl)quinolin-2- amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[4-chloranyl-3-(methylaminomethyl)phenoxy]methyl]quinolin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-08-20 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Phenyl Ether- and Aniline-Containing 2-Aminoquinolines as Potent and Selective Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
5A9I
 
 | | Crystal structure of the extracellular domain of PepT2 | | Descriptor: | CITRIC ACID, GLYCEROL, SOLUTE CARRIER FAMILY 15 MEMBER 2 | | Authors: | Beale, J.H, Newstead, S. | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal Structures of the Extracellular Domain from Pept1 and Pept2 Provide Novel Insights Into Mammalian Peptide Transport
Structure, 23, 2015
|
|
5B8A
 
 | | Crystal structure of oxidized chimeric EcAhpC1-186-YFSKHN | | Descriptor: | Alkyl hydroperoxide reductase subunit C,Peroxiredoxin-2, GLYCEROL, SULFATE ION | | Authors: | Kamariah, N, Sek, M.F, Eisenhaber, B, Eisenhaber, F, Gruber, G. | | Deposit date: | 2016-06-14 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Transition steps in peroxide reduction and a molecular switch for peroxide robustness of prokaryotic peroxiredoxins.
Sci Rep, 6, 2016
|
|
5BO6
 
 | | Structure of human sialyltransferase ST8SiaIII in complex with CDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Volkers, G, Worrall, L, Strynadka, N.C.J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of human ST8SiaIII sialyltransferase provides insight into cell-surface polysialylation.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8JIY
 
 | | A carbohydrate binding domain of a putative chondroitinase | | Descriptor: | DUF4955 domain-containing protein | | Authors: | Liu, G.C, Chang, Y.G. | | Deposit date: | 2023-05-29 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and structural characterization of a novel chondroitin sulfate-specific carbohydrate-binding module: The first member of a new family, CBM100.
Int.J.Biol.Macromol., 255, 2024
|
|
5BQQ
 
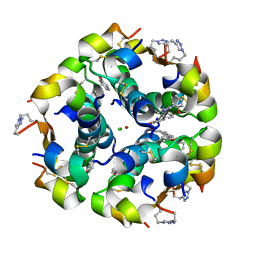 | | Human insulin with intra-chain chemical crosslink between modified B27 and B30 | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Brzozowski, A.M, Turkenburg, J.P, Jiracek, J, Zakova, L. | | Deposit date: | 2015-05-29 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Rational steering of insulin binding specificity by intra-chain chemical crosslinking.
Sci Rep, 6, 2016
|
|
8ITA
 
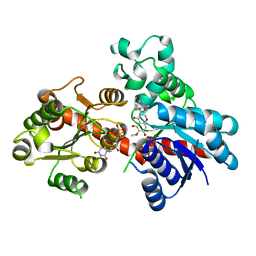 | | A reversible glycosyltransferase of tectorigenin - Bc7OUGT complexed with UDP and tectorigenin | | Descriptor: | 3-(4-hydroxyphenyl)-6-methoxy-5,7-bis(oxidanyl)chromen-4-one, Bc7OUGT, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhang, Z.Y, Lu, L, Guan, Z.F, Cheng, W.J. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional characterization and structural basis of a reversible glycosyltransferase involves in plant chemical defence.
Plant Biotechnol J, 21, 2023
|
|
5AM7
 
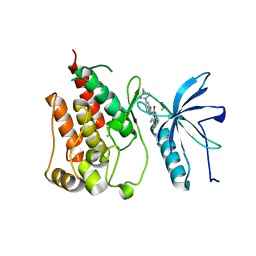 | | FGFR1 mutant with an inhibitor | | Descriptor: | 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, CHLORIDE ION, FIBROBLAST GROWTH FACTOR RECEPTOR 1 | | Authors: | Bunney, T.D, Wan, S, Thiyagarajan, N, Sutto, L, Williams, S.V, Ashford, P, Koss, H, Knowles, M.A, Gervasio, F.L, Coveney, P.V, Katan, M. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.957 Å) | | Cite: | The Effect of Mutations on Drug Sensitivity and Kinase Activity of Fibroblast Growth Factor Receptors: A Combined Experimental and Theoretical Study
Ebiomedicine, 2, 2015
|
|
5BN6
 
 | | Crystal Structure of Frutalin from Artocarpus incisa in complex with galactose | | Descriptor: | Jacalin, beta-D-galactopyranose | | Authors: | Vieira Neto, A.E, Pereira, H.M, Moreno, F.B.M.B, Moreira, A.C.O.M, Lobo, M.D.P, Sousa, F.D, Grangeiro, T.B, Moreira, R.A. | | Deposit date: | 2015-05-25 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6499 Å) | | Cite: | Crystal Structure of Frutalin from Artocarpus incisa in complex with galactose
To Be Published
|
|
5B0F
 
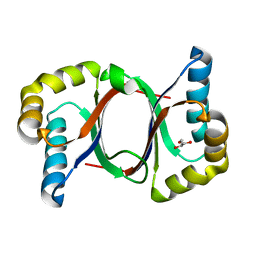 | | Polyketide cyclase OAC from Cannabis sativa, Y72F mutant | | Descriptor: | GLYCEROL, Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5BO9
 
 | | Structure of human sialyltransferase ST8SiaIII in complex with CMP-3FNeu5Ac and Sia-6S-LacNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Volkers, G, Worrall, L, Strynadka, N.C.J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human ST8SiaIII sialyltransferase provides insight into cell-surface polysialylation.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8J1C
 
 | |
8J1G
 
 | | Structure of amino acid dehydrogenase in complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Ohshima, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | First crystal structure of an NADP + -dependent l-arginine dehydrogenase belonging to the mu-crystallin family.
Int.J.Biol.Macromol., 249, 2023
|
|
5BPM
 
 | |
