4H3W
 
 | |
4HBR
 
 | |
2ZZF
 
 | | Crystal structure of alanyl-tRNA synthetase without oligomerization domain | | Descriptor: | Alanyl-tRNA synthetase, ZINC ION | | Authors: | Sokabe, M, Ose, T, Tokunaga, K, Nakamura, A, Nureki, O, Yao, M, Tanaka, I. | | Deposit date: | 2009-02-10 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of alanyl-tRNA synthetase with editing domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4HF7
 
 | |
2ZZT
 
 | | Crystal structure of the cytosolic domain of the cation diffusion facilitator family protein | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Higuchi, T, Hattori, M, Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2009-02-25 | | Release date: | 2009-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.842 Å) | | Cite: | Crystal structure of the cytosolic domain of the cation diffusion facilitator family protein
Proteins, 76, 2009
|
|
4HFQ
 
 | | Crystal structure of UDP-X diphosphatase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Duong-Ly, K.C, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2012-10-05 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A UDP-X diphosphatase from Streptococcus pneumoniae hydrolyzes precursors of peptidoglycan biosynthesis.
Plos One, 8, 2013
|
|
3A02
 
 | | Crystal structure of Aristaless homeodomain | | Descriptor: | CADMIUM ION, CHLORIDE ION, Homeobox protein aristaless | | Authors: | Miyazono, K, Nagata, K, Saigo, K, Kojima, T, Tanokura, M. | | Deposit date: | 2009-02-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Cooperative DNA-binding and sequence-recognition mechanism of aristaless and clawless
Embo J., 29, 2010
|
|
3A0V
 
 | | PAS domain of histidine kinase ThkA (TM1359) (SeMet, F486M/F489M) | | Descriptor: | ETHANOL, Sensor protein | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
4GOT
 
 | |
3A18
 
 | | Crystal Structure of Aldoxime Dehydratase (OxdRE) in Complex with Butyraldoxime (soaked crystal) | | Descriptor: | (1Z)-butanal oxime, Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sawai, H, Sugimoto, H, Kato, Y, Asano, Y, Shiro, Y, Aono, S. | | Deposit date: | 2009-03-26 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of michaelis complex of aldoxime dehydratase
J.Biol.Chem., 284, 2009
|
|
3A4U
 
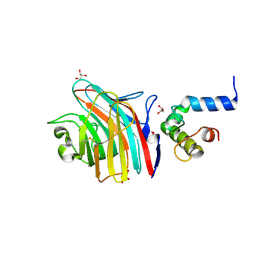 | | Crystal structure of MCFD2 in complex with carbohydrate recognition domain of ERGIC-53 | | Descriptor: | CALCIUM ION, GLYCEROL, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Nishio, M, Kamiya, Y, Mizushima, T, Wakatsuki, S, Sasakawa, H, Yamamoto, K, Uchiyama, S, Noda, M, McKay, A.R, Fukui, K, Hauri, H.P, Kato, K. | | Deposit date: | 2009-07-17 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the cooperative interplay between the two causative gene products of combined factor V and factor VIII deficiency.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3A8W
 
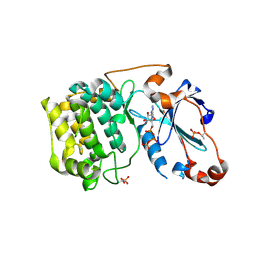 | | Crystal Structure of PKCiota kinase domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein kinase C iota type, SULFATE ION | | Authors: | Takimura, T, Kamata, K. | | Deposit date: | 2009-10-11 | | Release date: | 2010-05-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the PKC-iota kinase domain in its ATP-bound and apo forms reveal defined structures of residues 533-551 in the C-terminal tail and their roles in ATP binding
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4DSG
 
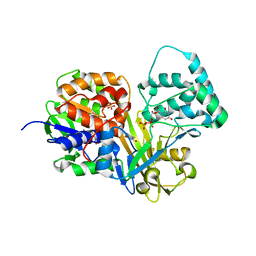 | | Crystal Structure of oxidized UDP-Galactopyranose mutase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, UDP-galactopyranose mutase, ... | | Authors: | Singh, H, Dhatwalia, R, Tanner, J.J. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Crystal Structures of Trypanosoma cruzi UDP-Galactopyranose Mutase Implicate Flexibility of the Histidine Loop in Enzyme Activation.
Biochemistry, 51, 2012
|
|
4E0P
 
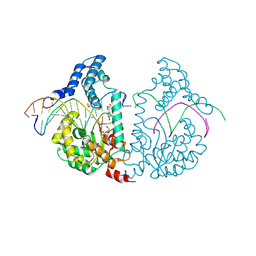 | | Protelomerase tela covalently complexed with substrate DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*TP*AP*AP*TP*AP*AP*CP*AP*AP*TP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*GP*AP*TP*AP*TP*TP*GP*TP*TP*AP*TP*TP*AP*TP*G)-3'), ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An enzyme-catalyzed multistep DNA refolding mechanism in hairpin telomere formation.
Plos Biol., 11, 2013
|
|
4E17
 
 | | Alpha-E-catenin is an autoinhibited molecule that co-activates vinculin | | Descriptor: | Catenin alpha-1, Vinculin | | Authors: | Choi, H.-J, Pokutta, S, Cadwell, G.W, Bankston, L.A, Liddington, R.C, Weis, W.I. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Conformational plasticity of alpha-catenin revealed by binding interactions with vinculin
To be Published
|
|
4E1L
 
 | | Crystal structure of Acetoacetyl-CoA thiolase (thlA2) from Clostridium difficile | | Descriptor: | Acetoacetyl-CoA thiolase 2, IODIDE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-06 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: |
|
|
4DW3
 
 | | Crystal structure of the glycoprotein Erns from the pestivirus BVDV-1 in complex with 5'-CMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Krey, T, Bontems, F, Vonrhein, C, Vaney, M.-C, Bricogne, G, Ruemenapf, T, Rey, F.A. | | Deposit date: | 2012-02-24 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Pestivirus Envelope Glycoprotein E(rns) and Mechanistic Analysis of Its Ribonuclease Activity.
Structure, 20, 2012
|
|
4DV1
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U20G, bound with streptomycin | | Descriptor: | 16S rRNA, MAGNESIUM ION, STREPTOMYCIN, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.849 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
4DWA
 
 | | Crystal structure of an active-site mutant of the glycoprotein Erns from the pestivirus BVDV-1 in complex with a CpUpC trinucleotide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, E(rns) glycoprotein, ... | | Authors: | Krey, T, Bontems, F, Vonrhein, C, Vaney, M.-C, Bricogne, G, Ruemenapf, T, Rey, F.A. | | Deposit date: | 2012-02-24 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structure of the Pestivirus Envelope Glycoprotein E(rns) and Mechanistic Analysis of Its Ribonuclease Activity.
Structure, 20, 2012
|
|
3A07
 
 | | Crystal Structure of Actinohivin; Potent anti-HIV Protein | | Descriptor: | Actinohivin, SODIUM ION | | Authors: | Tsunoda, M, Suzuki, K, Sagara, T, Takenaka, A. | | Deposit date: | 2009-03-04 | | Release date: | 2009-08-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Mechanism by which the lectin actinohivin blocks HIV infection of target cells
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4DWF
 
 | |
3AED
 
 | | Crystal structure of porcine heart mitochondrial complex II bound with 2-Iodo-N-phenyl-benzamide | | Descriptor: | 2-iodo-N-phenylbenzamide, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Sasaki, T, Shindo, M, Kido, Y, Inaoka, D.K, Omori, J, Osanai, A, Sakamoto, K, Mao, J, Matsuoka, S, Inoue, M, Honma, T, Tanaka, A, Kita, K. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal structure of porcine heart mitochondrial complex II bound with 2-Iodo-N-phenyl-benzamide
To be Published
|
|
4DXH
 
 | | Horse liver alcohol dehydrogenase complexed with NAD+ and 2,2,2-trifluoroethanol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alcohol dehydrogenase E chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), ... | | Authors: | Plapp, B.V, Ramaswamy, S. | | Deposit date: | 2012-02-27 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Atomic-Resolution Structures of Horse Liver Alcohol Dehydrogenase with NAD(+) and Fluoroalcohols Define Strained Michaelis Complexes.
Biochemistry, 51, 2012
|
|
4DYE
 
 | | Crystal structure of an enolase (putative sugar isomerase, target efi-502095) from streptomyces coelicolor, no mg, ordered loop | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, isomerase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of an enolase (putative sugar isomerase, target efi-502095) from streptomyces coelicolor, no mg, ordered loop
to be published
|
|
4DYZ
 
 | |
