2JA5
 
 | | CPD lesion containing RNA Polymerase II elongation complex A | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TTP*TP*TP*CP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | Deposit date: | 2006-11-23 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Cpd Damage Recognition by Transcribing RNA Polymerase II
Science, 315, 2007
|
|
5ITX
 
 | | Crystal Structure of Human NEIL1(P2G R242K) bound to duplex DNA containing Thymine Glycol | | Descriptor: | DNA (26-MER), Endonuclease 8-like 1 | | Authors: | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ITY
 
 | | Crystal Structure of Human NEIL1(P2G) bound to duplex DNA containing Thymine Glycol | | Descriptor: | DNA (26-MER), Endonuclease 8-like 1, GLYCEROL | | Authors: | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2JA8
 
 | | CPD lesion containing RNA Polymerase II elongation complex D | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TP*TP*TP*TTP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP *AP*GP*CP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*AP*UP)-3', ... | | Authors: | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | Deposit date: | 2006-11-23 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Cpd Damage Recognition by Transcribing RNA Polymerase II.
Science, 315, 2007
|
|
6K8P
 
 | |
3EIL
 
 | | Structure of B-DNA d(CGTTAATTAACG)2 in the presence of Manganese | | Descriptor: | 5'-D(*DCP*DGP*DTP*DTP*DAP*DAP*DTP*DTP*DAP*DAP*DCP*DG)-3', MANGANESE (II) ION | | Authors: | Millonig, H, Pous, J, Campos, L.J, Subirana, J.A. | | Deposit date: | 2008-09-16 | | Release date: | 2009-06-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interaction of manganese ions with DNA
J.Inorg.Biochem., 103, 2009
|
|
5VTP
 
 | |
2BTO
 
 | | Structure of BtubA from Prosthecobacter dejongeii | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, THIOREDOXIN 1, TUBULIN BTUBA | | Authors: | Schlieper, D, Lowe, J. | | Deposit date: | 2005-06-04 | | Release date: | 2005-06-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Bacterial Tubulin Btuba/B: Evidence for Horizontal Gene Transfer.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1EHL
 
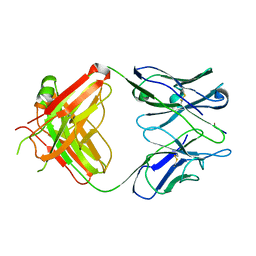 | | 64M-2 ANTIBODY FAB COMPLEXED WITH D(5HT)(6-4)T | | Descriptor: | 5'-(D(5HT)P*(6-4)T)-3', ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (HEAVY CHAIN), ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (LIGHT CHAIN) | | Authors: | Yokoyama, H, Mizutani, R, Satow, Y, Komatsu, Y, Ohtsuka, E, Nikaido, O. | | Deposit date: | 2000-02-21 | | Release date: | 2001-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the 64M-2 antibody Fab fragment in complex with a DNA dT(6-4)T photoproduct formed by ultraviolet radiation.
J.Mol.Biol., 299, 2000
|
|
1B3T
 
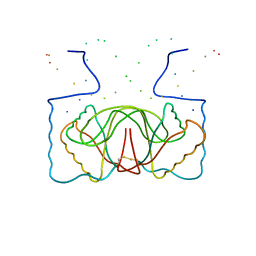 | | EBNA-1 NUCLEAR PROTEIN/DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*GP*GP*AP*AP*GP*CP*AP*TP*AP*TP*GP*CP*TP*TP*CP*CP*C)-3'), PROTEIN (NUCLEAR PROTEIN EBNA1) | | Authors: | Bochkarev, A, Bochkareva, E, Edwards, A, Frappier, L. | | Deposit date: | 1998-12-14 | | Release date: | 1998-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A structure of a permanganate-sensitive DNA site bound by the Epstein-Barr virus origin binding protein, EBNA1.
J.Mol.Biol., 284, 1998
|
|
4A08
 
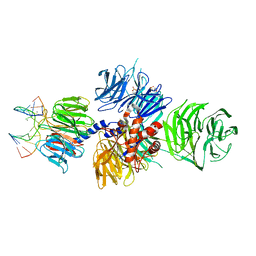 | | Structure of hsDDB1-drDDB2 bound to a 13 bp CPD-duplex (purine at D-1 position) at 3.0 A resolution (CPD 1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*AP*CP*GP*CP*GP*AP*(TTD)P*GP*CP*GP*CP*CP*C)-3', 5'-D(*TP*GP*GP*GP*CP*GP*CP*CP*CP*TP*CP*GP*CP*G)-3', ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
5Z8F
 
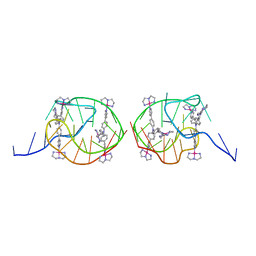 | | Solution structure for the unique dimeric 4:2 complex of a platinum(II)-based tripod bound to a hybrid-1 human telomeric G-quadruplex | | Descriptor: | 4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]-N,N-bis[4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]phenyl]aniline, G-quadruplex DNA (26-MER) | | Authors: | Liu, W.T, Zhong, Y.F, Liu, L.Y, Zeng, W.J, Wang, F.Y, Yang, D.Z, Mao, Z.W. | | Deposit date: | 2018-01-31 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding
Nat Commun, 9, 2018
|
|
1RRU
 
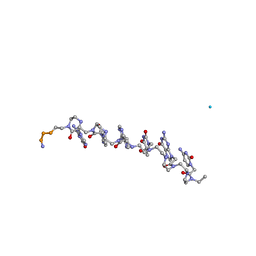 | | The influence of a chiral amino acid on the helical handedness of PNA in solution and in crystals | | Descriptor: | Peptide Nucleic Acid, (H-P(*CPN*GPN*TPN*APN*CPN*GPN)-LYS-NH2) | | Authors: | Rasmussen, H, Liljefors, T, Petersson, B, Nielsen, P.E, Kastrup, J.S. | | Deposit date: | 2003-12-09 | | Release date: | 2004-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Influence of a Chiral Amino Acid on the Helical Handedness of PNA in Solution and in Crystals
J.Biomol.Struct.Dyn., 21, 2004
|
|
4A0A
 
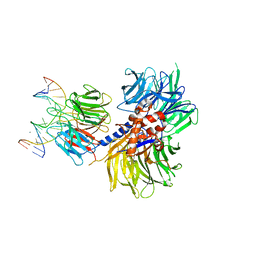 | | Structure of hsDDB1-drDDB2 bound to a 16 bp CPD-duplex (pyrimidine at D-1 position) at 3.6 A resolution (CPD 3) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*GP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*DGP)-3', CALCIUM ION, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4A09
 
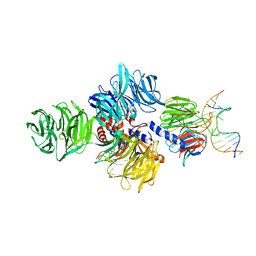 | | Structure of hsDDB1-drDDB2 bound to a 15 bp CPD-duplex (purine at D-1 position) at 3.1 A resolution (CPD 2) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*GP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*GP)-3', CALCIUM ION, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0B
 
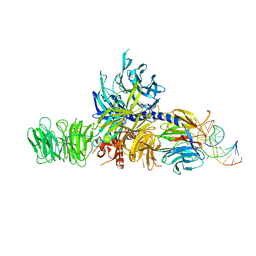 | | Structure of hsDDB1-drDDB2 bound to a 16 bp CPD-duplex (pyrimidine at D-1 position) at 3.8 A resolution (CPD 4) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*DGP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*DGP)-3', DNA DAMAGE-BINDING PROTEIN 1, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
2XX3
 
 | |
1M69
 
 | |
4HN1
 
 | | Crystal Structure of H60N/Y130F double mutant of ChmJ, a 3'-monoepimerase from Streptomyces bikiniensis in complex with dTDP | | Descriptor: | 1,2-ETHANEDIOL, Putative 3-epimerase in D-allose pathway, THYMIDINE, ... | | Authors: | Holden, H.M, Kubiak, R.L. | | Deposit date: | 2012-10-18 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Studies on a 3'-Epimerase Involved in the Biosynthesis of dTDP-6-deoxy-d-allose.
Biochemistry, 51, 2012
|
|
7YC7
 
 | | Dark, fully reduced structure of the MmCPDII-DNA complex as produced at SwissFEL | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
2XRZ
 
 | | X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei in complex with intact CPD-lesion | | Descriptor: | ACETATE ION, COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, ... | | Authors: | Kiontke, S, Geisselbrecht, Y, Pokorny, R, Carell, T, Batschauer, A, Essen, L.O. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of an Archaeal Class II DNA Photolyase and its Complex with Uv-Damaged Duplex DNA.
Embo J., 30, 2011
|
|
7YD7
 
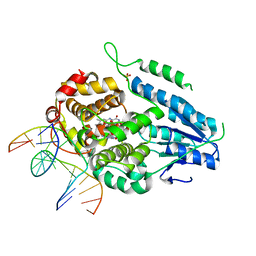 | | TR-SFX MmCPDII-DNA complex: 1 ns snapshot. Includes 1 ns, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YCM
 
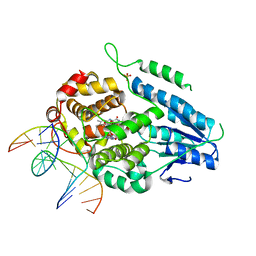 | | TR-SFX MmCPDII-DNA complex: 100 ps snapshot. Includes 100ps, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YCR
 
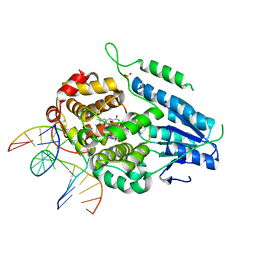 | | TR-SFX MmCPDII-DNA complex: 450 ps snapshot. Includes 450ps, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YCP
 
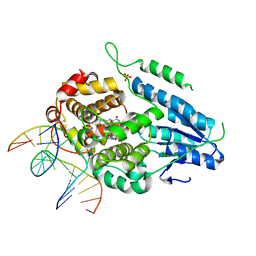 | | TR-SFX MmCPDII-DNA complex: 250 ps snapshot. Includes 250 ps, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
