2I06
 
 | | Escherichia Coli Replication Terminator Protein (Tus) Complexed With DNA- Locked form | | Descriptor: | 5'-D(*T*TP*AP*GP*TP*TP*AP*CP*AP*AP*CP*AP*TP*AP*CP*T)-3', 5'-D(*TP*G*AP*TP*AP*TP*GP*TP*TP*GP*TP*AP*AP*CP*TP*A)-3', DNA replication terminus site-binding protein, ... | | Authors: | Oakley, A.J, Mulcair, M.D, Schaeffer, P.M, Dixon, N.E. | | Deposit date: | 2006-08-10 | | Release date: | 2007-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Polarity of Termination of DNA Replication in E. coli.
To be Published
|
|
2I05
 
 | | Escherichia Coli Replication Terminator Protein (Tus) Complexed With TerA DNA | | Descriptor: | 5'-D(*T*TP*AP*GP*TP*TP*AP*CP*AP*AP*CP*AP*TP*AP*CP*T)-3', 5'-D(*TP*AP*GP*TP*AP*TP*GP*TP*TP*GP*TP*AP*AP*CP*TP*A)-3', DNA replication terminus site-binding protein, ... | | Authors: | Oakley, A.J, Mulcair, M.D, Schaeffer, P.M, Dixon, N.E. | | Deposit date: | 2006-08-10 | | Release date: | 2007-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Polarity of Termination of DNA Replication in E. coli
To be Published
|
|
1KE6
 
 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH N-METHYL-{4-[2-(7-OXO-6,7-DIHYDRO-8H-[1,3]THIAZOLO[5,4-E]INDOL-8-YLIDENE)HYDRAZINO]PHENYL}METHANESULFONAMIDE | | Descriptor: | Cell division protein kinase 2, N-METHYL-{4-[2-(7-OXO-6,7-DIHYDRO-8H-[1,3]THIAZOLO[5,4-E]INDOL-8-YLIDENE)HYDRAZINO]PHENYL}METHANESULFONAMIDE | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.H, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
5UFM
 
 | | Crystal structure of Burkholderia thailandensis 1,6-didemethyltoxoflavin-N1-methyltransferase with bound 1,6-didemethyltoxoflavin and S-adenosylhomocysteine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Methyltransferase domain protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Fenwick, M.K, Ealick, S.E, Almabruk, K.H, Begley, T.P, Philmus, B. | | Deposit date: | 2017-01-05 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Biochemical Characterization and Structural Basis of Reactivity and Regioselectivity Differences between Burkholderia thailandensis and Burkholderia glumae 1,6-Didesmethyltoxoflavin N-Methyltransferase.
Biochemistry, 56, 2017
|
|
4O4K
 
 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{E})-2-azanylidene-5-[(4-hydroxyphenyl)methylidene]-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
4MKB
 
 | |
1I08
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE H30A MUTANT OF MANGANESE SUPEROXIDE DISMUTASE FROM E. COLI | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Baker, E.N, Jameson, G.B. | | Deposit date: | 2001-01-29 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Removing a hydrogen bond in the dimer interface of Escherichia coli manganese superoxide dismutase alters structure and reactivity.
Biochemistry, 40, 2001
|
|
1I0H
 
 | | CRYSTAL STRUCTURE OF THE E. COLI MANGANESE SUPEROXIDE DISMUTASE MUTANT Y174F AT 1.35 ANGSTROMS RESOLUTION. | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE Y174F MUTANT | | Authors: | Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Baker, E.N, Jameson, G.B. | | Deposit date: | 2001-01-29 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Removing a hydrogen bond in the dimer interface of Escherichia coli manganese superoxide dismutase alters structure and reactivity.
Biochemistry, 40, 2001
|
|
3GOL
 
 | | HCV NS5b polymerase in complex with 1,5 benzodiazepine inhibitor (R)-11d | | Descriptor: | (11R)-10-acetyl-11-(2,4-dichlorophenyl)-6-hydroxy-3,3-dimethyl-2,3,4,5,10,11-hexahydro-1H-dibenzo[b,e][1,4]diazepin-1-one, MAGNESIUM ION, RNA-directed RNA polymerase | | Authors: | Nyanguile, O, De Bondt, H. | | Deposit date: | 2009-03-19 | | Release date: | 2009-06-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 1,5-Benzodiazepine inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4O43
 
 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{E})-3-[(2~{R})-butan-2-yl]-5-[(4-hydroxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
4O5G
 
 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{E})-5-[(4-aminophenyl)methylidene]-2-azanylidene-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
4G1Q
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with Rilpivirine (TMC278, Edurant), a non-nucleoside rt-inhibiting drug | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, MAGNESIUM ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2012-07-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Snapshot of the equilibrium dynamics of a drug bound to HIV-1 reverse transcriptase.
Nat Chem, 5, 2013
|
|
4LRG
 
 | | Structure of BRD4 bromodomain 1 with a dimethyl thiophene isoxazole azepine carboxamide | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-[1,2]oxazolo[5,4-c]thieno[2,3-e]azepin-6-yl]acetamide, Bromodomain-containing protein 4 | | Authors: | Ravichandran, S, Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
5QIW
 
 | | Covalent fragment group deposition -- Crystal Structure of OUTB2 in complex with PCM-0102660 | | Descriptor: | N-[(E)-(4-methylphenyl)methylidene]acetamide, UNKNOWN LIGAND, Ubiquitin thioesterase OTUB2 | | Authors: | Sethi, R, Douangamath, A, Resnick, E, Bradley, A.R, Collins, P, Brandao-Neto, J, Talon, R, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, London, N, von Delft, F. | | Deposit date: | 2018-08-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Covalent fragment group deposition
To Be Published
|
|
5QIT
 
 | | Covalent fragment group deposition -- Crystal Structure of OUTB2 in complex with PCM-0102821 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, N-[(E)-(3-methylphenyl)methylidene]acetamide, ... | | Authors: | Sethi, R, Douangamath, A, Resnick, E, Bradley, A.R, Collins, P, Brandao-Neto, J, Talon, R, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, London, N, von Delft, F. | | Deposit date: | 2018-08-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Covalent fragment group deposition
To Be Published
|
|
1S1X
 
 | | Crystal structure of V108I mutant HIV-1 reverse transcriptase in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
1QZM
 
 | | alpha-domain of ATPase | | Descriptor: | ATP-dependent protease La | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Khalatova, A.G, Rasulova, F.S, Tropea, J.E, Maurizi, M.R, Rotanova, T.V, Gustchina, A, Wlodawer, A. | | Deposit date: | 2003-09-17 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the AAA+ alpha domain of E. coli Lon protease at 1.9A resolution.
J.Struct.Biol., 146
|
|
1S1U
 
 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
2GBH
 
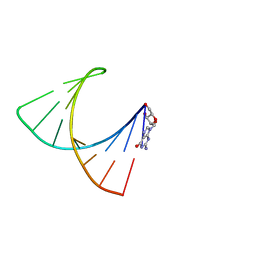 | | NMR structure of stem region of helix-35 of 23S E.coli ribosomal RNA (residues 736-760) | | Descriptor: | 5'-R(*(GMP)P*GP*GP*CP*UP*AP*AP*UP*GP*(PSU)P*UP*GP*AP*AP*AP*AP*AP*UP*UP*AP*GP*CP*CP*C)-3' | | Authors: | Bax, A, Boisbouvier, J, Bryce, D, Grishaev, A, Jaroniec, C, Miclet, E, Nikonovicz, E, O'Neil-Cabello, E, Ying, J. | | Deposit date: | 2006-03-10 | | Release date: | 2006-04-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Measurement of five dipolar couplings from a single 3D NMR multiplet applied to the study of RNA dynamics.
J.Am.Chem.Soc., 126, 2004
|
|
2YGY
 
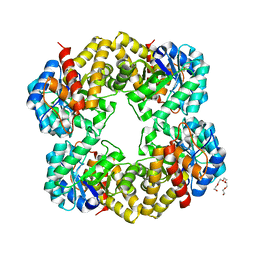 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in space group P21 crystal form II | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL | | Authors: | Campeotto, I, Nelson, A, Berry, A, Phillips, S.E.V, Pearson, A.R. | | Deposit date: | 2011-04-23 | | Release date: | 2012-04-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pathological macromolecular crystallographic data affected by twinning, partial-disorder and exhibiting multiple lattices for testing of data processing and refinement tools.
Sci Rep, 8, 2018
|
|
1G15
 
 | |
1G1T
 
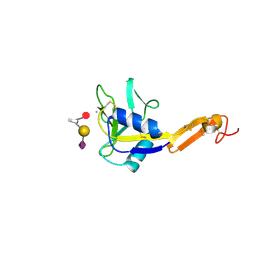 | | CRYSTAL STRUCTURE OF E-SELECTIN LECTIN/EGF DOMAINS COMPLEXED WITH SLEX | | Descriptor: | CALCIUM ION, E-SELECTIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Somers, W.S, Camphausen, R.T. | | Deposit date: | 2000-10-13 | | Release date: | 2001-10-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1.
Cell(Cambridge,Mass.), 103, 2000
|
|
1PCL
 
 | |
6I1S
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with FKBP12 and the inhibitor E6201 | | Descriptor: | (4~{S},5~{R},6~{Z},9~{S},10~{S},12~{E})-16-(ethylamino)-4,5-dimethyl-9,10,18-tris(oxidanyl)-3-oxabicyclo[12.4.0]octadeca-1(14),6,12,15,17-pentaene-2,8-dione, 1,2-ETHANEDIOL, Activin receptor type-1, ... | | Authors: | Williams, E.P, Pinkas, D.M, Fortin, J, Newman, J.A, Bradshaw, W.J, Mahajan, P, Kupinska, K, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-10-30 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Mutant ACVR1 Arrests Glial Cell Differentiation to Drive Tumorigenesis in Pediatric Gliomas.
Cancer Cell, 37, 2020
|
|
