8BZB
 
 | | co-soak stabilizers for ERa - 14-3-3 interaction (884_AZ275) | | Descriptor: | 14-3-3 protein sigma, 2-(4-chloranylphenoxy)-2-methyl-~{N}-(2-sulfanylethyl)propanamide, 4-ethoxy-1-benzothiophene-2-carboximidamide, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BZ0
 
 | | single soak stabilizer for ERa - 14-3-3 interaction (AZ275) | | Descriptor: | 14-3-3 protein sigma, 4-ethoxy-1-benzothiophene-2-carboximidamide, ERalpha peptide, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8C4G
 
 | | Small molecule amidine soak in 14-3-3/ERa (AZ132) | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, MAGNESIUM ION, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2023-01-03 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3D48
 
 | |
4P8Q
 
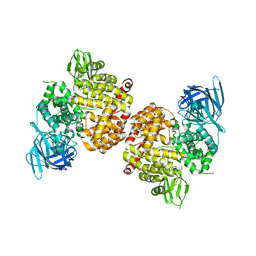 | | Crystal Structure of Human Insulin Regulated Aminopeptidase with Alanine in Active Site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucyl-cystinyl aminopeptidase, ... | | Authors: | Hermans, S.J, Ascher, D.B, Hancock, N.C, Holien, J.K, Michell, B, Morton, C.J, Parker, M.W. | | Deposit date: | 2014-03-31 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Crystal structure of human insulin-regulated aminopeptidase with specificity for cyclic peptides.
Protein Sci., 24, 2015
|
|
8BZA
 
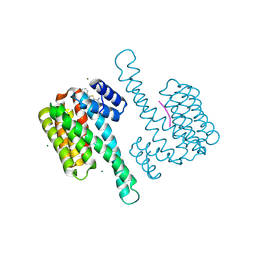 | | single soak stabilizer for ERa - 14-3-3 interaction (AZ555) | | Descriptor: | 14-3-3 protein sigma, 4-methyl-5-phenyl-thiophene-2-carboximidamide, ERalpha peptide, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CRH
 
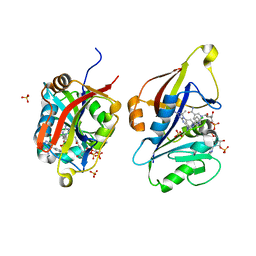 | | Crystal structure of Candida auris dihydrofolate reductase complexed with NADPH and cycloguanil | | Descriptor: | 5-(4-chlorophenyl)-6,6-dimethyl-1,4-dihydro-1,3,5-triazine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kirkman, T.J, Dias, M.V.B. | | Deposit date: | 2023-03-08 | | Release date: | 2023-08-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of dihydrofolate reductase from the emerging pathogenic fungus Candida auris.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8IM0
 
 | | mCherry-LaM8 complex | | Descriptor: | LaM8, MCherry fluorescent protein | | Authors: | Liang, H, Liu, R, Ding, Y. | | Deposit date: | 2023-03-05 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural Insights into the Binding of Red Fluorescent Protein mCherry-Specific Nanobodies.
Int J Mol Sci, 24, 2023
|
|
8C0K
 
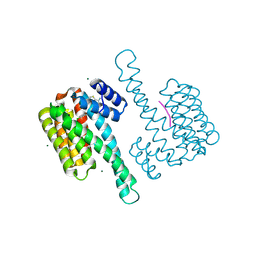 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (1075302) | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, MAGNESIUM ION, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-12-17 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BYY
 
 | |
3LXO
 
 | | The crystal structure of ribonuclease A in complex with thymidine-3'-monophosphate | | Descriptor: | Ribonuclease pancreatic, THYMIDINE-3'-PHOSPHATE | | Authors: | Doucet, N, Jayasundera, T.B, Simonovic, M, Loria, J.P. | | Deposit date: | 2010-02-25 | | Release date: | 2010-04-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | The crystal structure of ribonuclease A in complex with thymidine-3'-monophosphate provides further insight into ligand binding.
Proteins, 78, 2010
|
|
8C04
 
 | | Co-soaked stabilizers for ERa - 14-3-3 interaction (884_AZ354) | | Descriptor: | 14-3-3 protein sigma, 2-(4-chloranylphenoxy)-2-methyl-~{N}-(2-sulfanylethyl)propanamide, 4-chloranyl-7-propan-2-yloxy-1-benzothiophene-2-carboximidamide, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2022-12-15 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8C4F
 
 | | Small molecule amidine soak in 14-3-3/ERa (AZ037) | | Descriptor: | 14-3-3 protein sigma, 5-(cyclohexylamino)-4-phenyl-thiophene-2-carboximidamide, ERalpha peptide, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2023-01-03 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8IDN
 
 | | Cryo-EM structure of RBD/E77-Fab complex | | Descriptor: | E77 Fab heavy chain, E77 Fab light chain, Spike protein S1, ... | | Authors: | Lu, D.F, Zhang, Z.C. | | Deposit date: | 2023-02-13 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | The structure of the RBD-E77 Fab complex reveals neutralization and immune escape of SARS-CoV-2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
3CL3
 
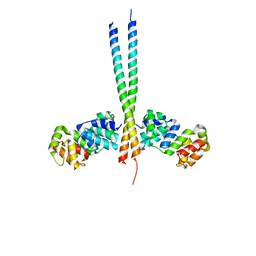 | | Crystal Structure of a vFLIP-IKKgamma complex: Insights into viral activation of the IKK signalosome | | Descriptor: | NF-kappa-B essential modulator, ORF K13 | | Authors: | Bagneris, C, Ageichik, A.V, Cronin, N, Boshoff, C, Waksman, G, Barrett, T. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a vFlip-IKKgamma complex: insights into viral activation of the IKK signalosome.
Mol.Cell, 30, 2008
|
|
3D69
 
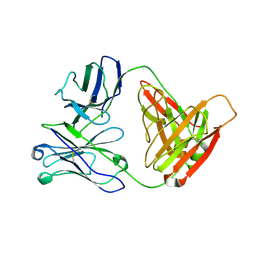 | |
4PC6
 
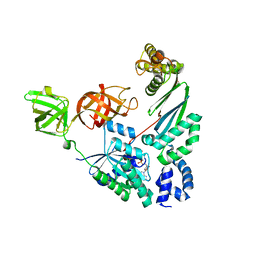 | | Elongation factor Tu:Ts complex with bound GDPNP | | Descriptor: | Elongation factor Ts, Elongation factor Tu, GLYCEROL, ... | | Authors: | Thirup, S.S. | | Deposit date: | 2014-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu.
J.Struct.Biol., 191, 2015
|
|
4PH7
 
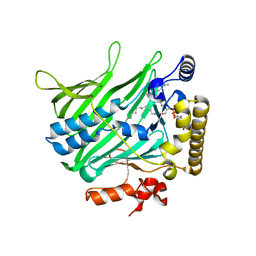 | | Structure of Osh6p in complex with phosphatidylinositol 4-phosphate | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, Oxysterol-binding protein homolog 6 | | Authors: | Delfosse, V, Moser von Filseck, J, Antonny, B, Bourguet, W, Drin, G. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | INTRACELLULAR TRANSPORT. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate.
Science, 349, 2015
|
|
7NM3
 
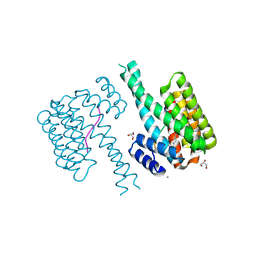 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-135 | | Descriptor: | 14-3-3 protein sigma, 4-[4-(dimethylamino)piperidin-1-yl]sulfonylbenzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
3D7E
 
 | |
7NK5
 
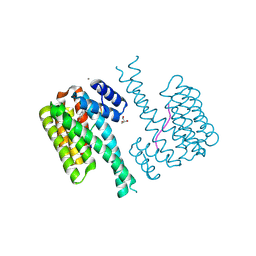 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-124 | | Descriptor: | 1-(4-methylphenyl)sulfonyl-4-(2-methylpropyl)piperazine, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-17 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
8BYZ
 
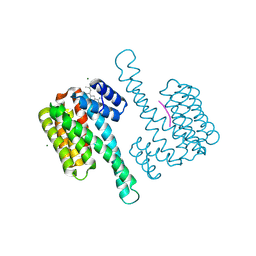 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (AZ210) | | Descriptor: | 14-3-3 protein sigma, 4-[(2~{S})-3-azanyl-2-methyl-propyl]-7-methoxy-1-benzothiophene-2-carboximidamide, ERalpha peptide, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5AP9
 
 | | Controlled lid-opening in Thermomyces lanuginosus lipase - a switch for activity and binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Skjold-Joergensen, J, Vind, J, Moroz, O.V, Blagova, E.V, Bhatia, V.K, Svendsen, A, Wilson, K.S, Bjerrum, M.J. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Controlled lid-opening in Thermomyces lanuginosus lipase- An engineered switch for studying lipase function.
Biochim. Biophys. Acta, 1865, 2017
|
|
7S9Z
 
 | | Helicobacter Hepaticus CcsBA Closed Conformation | | Descriptor: | Cytochrome c biogenesis protein, HEME B/C, PHOSPHATIDYLETHANOLAMINE | | Authors: | Mendez, D.L, Lowder, E.P, Tillman, D.E, Sutherland, M.C, Collier, A.L, Rau, M.J, Fitzpatrick, J.A, Kranz, R.G. | | Deposit date: | 2021-09-21 | | Release date: | 2021-12-22 | | Last modified: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Cryo-EM of CcsBA reveals the basis for cytochrome c biogenesis and heme transport.
Nat.Chem.Biol., 18, 2022
|
|
7NLE
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-118 | | Descriptor: | 14-3-3 protein sigma, 4-(4-methylpiperazin-1-yl)sulfonylbenzaldehyde, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
