5AEZ
 
 | |
5LHA
 
 | |
8I3O
 
 | |
8I4D
 
 | | X-ray structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex at 100K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yano, N, Kondo, T, Kusaka, K, Yamada, T, Arakawa, T, Sakamoto, T, Fushinobu, S. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Charge neutralization and beta-elimination cleavage mechanism of family 42 L-rhamnose-alpha-1,4-D-glucuronate lyase revealed using neutron crystallography.
J.Biol.Chem., 300, 2024
|
|
8IDZ
 
 | |
5LGV
 
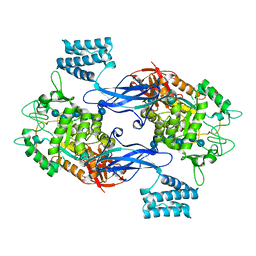 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant soaked in maltooctaose | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Mia, F, Barclay, J.E, Tang, M, Gorelik, A, Rashid, A.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2016-07-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-bound structures and site-directed mutagenesis identify the acceptor and secondary binding sites of Streptomyces coelicolor maltosyltransferase GlgE.
J.Biol.Chem., 291, 2016
|
|
8EVZ
 
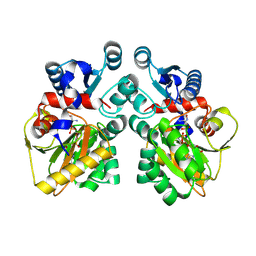 | | DdlB from Pseudomonas aeruginosa PAO1 in complex with ADP and phosphorylated D-cycloserine | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine--D-alanine ligase B, MAGNESIUM ION, ... | | Authors: | Pederick, J.L, Woolman, J.C, Bruning, J.B. | | Deposit date: | 2022-10-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Comparative functional and structural analysis of Pseudomonas aeruginosa d-alanine-d-alanine ligase isoforms as prospective antibiotic targets.
Febs J., 290, 2023
|
|
5AHE
 
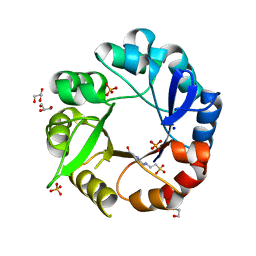 | | Crystal structure of Salmonella enterica HisA | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Soderholm, A, Guo, X, Newton, M.S, Evans, G.B, Nasvall, J, Patrick, W.M, Selmer, M. | | Deposit date: | 2015-02-05 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two-Step Ligand Binding in a Beta/Alpha8 Barrel Enzyme -Substrate-Bound Structures Shed New Light on the Catalytic Cycle of Hisa
J.Biol.Chem., 290, 2015
|
|
8I3P
 
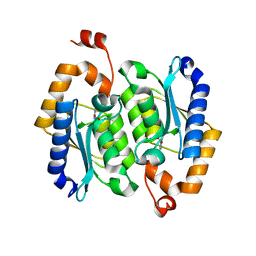 | | crystal structure of yeast cytosine deaminase mutant yCD-RQ-1/8SAH in complex with (R)-4-hydroxy-3,4-dihydropyrimidin-2(1H)-one | | Descriptor: | 1,2-ETHANEDIOL, 4-oxidanyl-3,4-dihydro-1H-pyrimidin-2-one, Cytosine deaminase, ... | | Authors: | Qin, M.M, Deng, H.Z, Yao, L.S. | | Deposit date: | 2023-01-17 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | crystal structure of yeast cytosine deaminase mutant yCD-RQ-1/8SAH in complex with (R)-4-hydroxy-3,4-dihydropyrimidin-2(1H)-one
To be published
|
|
6X1N
 
 | |
5ADZ
 
 | | Ether Lipid-Generating Enzyme AGPS in complex with inhibitor 1a | | Descriptor: | (3S)-3-(2-fluorophenyl)-N-((2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)methyl)butanamide), ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, PEROXISOMAL, ... | | Authors: | Piano, V, Benjamin, D.I, Valente, S, Nenci, S, Marrocco, B, Mai, A, Aliverti, A, Nomura, D.K, Mattevi, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Inhibitors for the Ether Lipid-Generating Enzyme Agps as Anti-Cancer Agents.
Acs Chem.Biol., 10, 2015
|
|
8EVW
 
 | | DdlA from Pseudomonas aeruginosa PAO1 in complex with ATP and D-ala-D-ala | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-ALANINE, D-alanine--D-alanine ligase A, ... | | Authors: | Pederick, J.L, Woolman, J.C, Bruning, J.B. | | Deposit date: | 2022-10-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Comparative functional and structural analysis of Pseudomonas aeruginosa d-alanine-d-alanine ligase isoforms as prospective antibiotic targets.
Febs J., 290, 2023
|
|
5AHS
 
 | | 3-Sulfinopropionyl-Coenzyme A (3SP-CoA) desulfinase from Advenella mimgardefordensis DPN7T: holo crystal structure with the substrate analog succinyl-CoA | | Descriptor: | ACYL-COA DEHYDROGENASE, COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cianci, M, Schuermann, M, Meijers, R, Schneider, T.R, Steinbuechel, A. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Sulfinopropionyl-Coenzyme a (3Sp-Coa) Desulfinase from Advenella Mimigardefordensis Dpn7(T): Crystal Structure and Function of a Desulfinase with an Acyl-Coa Dehydrogenase Fold.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8I3N
 
 | |
4ZSL
 
 | | MITOGEN ACTIVATED PROTEIN KINASE 7 IN COMPLEX WITH INHIBITOR | | Descriptor: | 3-amino-5-[(4-chlorophenyl)amino]-N-[(1S)-1-phenylethyl]-1H-1,2,4-triazole-1-carboxamide, GLYCEROL, Mitogen-activated protein kinase 7 | | Authors: | Ogg, D.J, Tucker, J. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8EJ0
 
 | | Crystal structure of Fe-S cluster-dependent dehydratase from Paralcaligenes ureilyticus in complex with Mg | | Descriptor: | BICARBONATE ION, CARBON DIOXIDE, Dihydroxyacid dehydratase, ... | | Authors: | Bayaraa, T, Lonhienne, T, Guddat, L.W. | | Deposit date: | 2022-09-16 | | Release date: | 2023-08-30 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural and Functional Insight into the Mechanism of the Fe-S Cluster-Dependent Dehydratase from Paralcaligenes ureilyticus.
Chemistry, 29, 2023
|
|
8EVV
 
 | |
5LFQ
 
 | | Crystal Structure of the Bacterial Proteasome Activator Bpa of Mycobacterium tuberculosis (space group P3) | | Descriptor: | Bacterial proteasome activator | | Authors: | Bolten, M, Delley, C.L, Leibundgut, M, Boehringer, D, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structural Analysis of the Bacterial Proteasome Activator Bpa in Complex with the 20S Proteasome.
Structure, 24, 2016
|
|
8EVX
 
 | | DdlA from Pseudomonas aeruginosa PAO1 in complex with ADP and phosphorylated D-cycloserine | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine--D-alanine ligase A, MAGNESIUM ION, ... | | Authors: | Pederick, J.L, Woolman, J.C, Bruning, J.B. | | Deposit date: | 2022-10-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Comparative functional and structural analysis of Pseudomonas aeruginosa d-alanine-d-alanine ligase isoforms as prospective antibiotic targets.
Febs J., 290, 2023
|
|
5LGT
 
 | | Thieno[3,2-b]pyrrole-5-carboxamides as Novel Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1: Compound 15 | | Descriptor: | 4-methyl-~{N}-[2-[[4-(1-methylpiperidin-4-yl)oxyphenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, ... | | Authors: | Mattevi, A, Ciossani, G. | | Deposit date: | 2016-07-08 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
4ZT5
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor (2S)-N-(3,5-dichlorobenzyl)-N'-(1H-imidazo[4,5-b]pyridin-2-yl)-2-methylpropane-1,3-diamine (Chem 1655) | | Descriptor: | (2S)-N-(3,5-dichlorobenzyl)-N'-(1H-imidazo[4,5-b]pyridin-2-yl)-2-methylpropane-1,3-diamine, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.-Y, Hol, W.G.J. | | Deposit date: | 2015-05-14 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 5-Fluoroimidazo[4,5-b]pyridine Is a Privileged Fragment That Conveys Bioavailability to Potent Trypanosomal Methionyl-tRNA Synthetase Inhibitors.
Acs Infect Dis., 2, 2016
|
|
6WOJ
 
 | | Structure of the SARS-CoV-2 macrodomain (NSP3) in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Gao, F.P, Fehr, A.R. | | Deposit date: | 2020-04-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The SARS-CoV-2 Conserved Macrodomain Is a Mono-ADP-Ribosylhydrolase.
J.Virol., 95, 2021
|
|
5LK0
 
 | |
6WH9
 
 | | Ketoreductase from module 1 of the 6-deoxyerythronolide B synthase (KR1) in complex with antibody fragment (Fab) 1D10 | | Descriptor: | 1D10 (Fab heavy chain), 1D10 (Fab light chain), KR1, ... | | Authors: | Cogan, D.P, Mathews, I.I, Khosla, C. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Antibody Probes of Module 1 of the 6-Deoxyerythronolide B Synthase Reveal an Extended Conformation During Ketoreduction.
J.Am.Chem.Soc., 142, 2020
|
|
5LKQ
 
 | | Protease domain of RadA | | Descriptor: | DNA repair protein RadA | | Authors: | Rapisarda, C, Fronzes, R. | | Deposit date: | 2016-07-22 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Bacterial RadA is a DnaB-type helicase interacting with RecA to promote bidirectional D-loop extension.
Nat Commun, 8, 2017
|
|
