8EHG
 
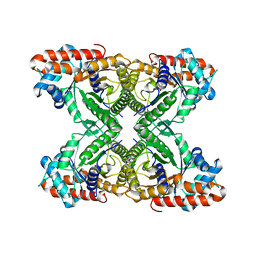 | | Rabbit muscle aldolase determined using single-particle cryo-EM with Apollo camera. | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Peng, R, Fu, X, Mendez, J.H, Randolph, P.H, Bammes, B, Stagg, S.M. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Characterizing the resolution and throughput of the Apollo direct electron detector.
J Struct Biol X, 7, 2023
|
|
8EMQ
 
 | | Mouse apoferritin heavy chain with zinc determined using single-particle cryo-EM with Apollo camera. | | Descriptor: | FE (III) ION, Ferritin heavy chain, N-terminally processed, ... | | Authors: | Peng, R, Fu, X, Mendez, J.H, Randolph, P.H, Bammes, B, Stagg, S.M. | | Deposit date: | 2022-09-28 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (1.66 Å) | | Cite: | Characterizing the resolution and throughput of the Apollo direct electron detector.
J Struct Biol X, 7, 2023
|
|
8EN7
 
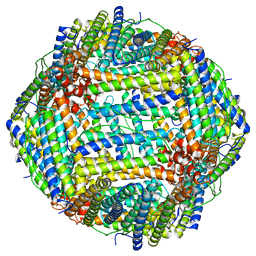 | | Mouse apoferritin heavy chain without zinc determined using single-particle cryo-EM with Apollo camera. | | Descriptor: | FE (III) ION, Ferritin heavy chain, N-terminally processed | | Authors: | Peng, R, Fu, X, Mendez, J.H, Randolph, P.H, Bammes, B, Stagg, S.M. | | Deposit date: | 2022-09-28 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (1.68 Å) | | Cite: | Characterizing the resolution and throughput of the Apollo direct electron detector.
J Struct Biol X, 7, 2023
|
|
6F8T
 
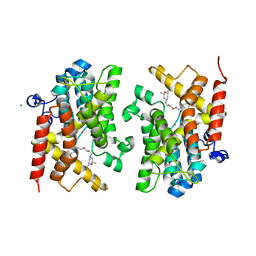 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-4a | | Descriptor: | (2~{R})-1-[(~{E})-(3-cyclopentyloxy-4-methoxy-phenyl)methylideneamino]oxy-3-[(2~{R},6~{S})-2,6-dimethylmorpholin-4-yl]propan-2-ol, MAGNESIUM ION, ZINC ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6F8V
 
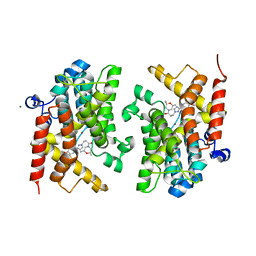 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-18b | | Descriptor: | 3-[3-(3-cyclopentyloxy-4-methoxy-phenyl)pyrazol-1-yl]-1-[(2~{R},6~{R})-2,6-dimethylmorpholin-4-yl]propan-1-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6F8U
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-20b | | Descriptor: | 2-[(~{E})-[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]methylideneamino]oxy-1-[(2~{R},6~{R})-2,6-dimethylmorpholin-4-yl]ethanone, MAGNESIUM ION, ZINC ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6F8X
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-26g | | Descriptor: | 1,2-ETHANEDIOL, 2-[(5~{R})-3-(3-cyclopentyloxy-4-methoxy-phenyl)-4,5-dihydro-1,2-oxazol-5-yl]-~{N},~{N}-bis(2-hydroxyethyl)ethanamide, DIMETHYL SULFOXIDE, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6F8R
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-54 | | Descriptor: | (2~{S})-1-[3-(3-cyclopentyloxy-4-methoxy-phenyl)pyrazol-1-yl]-3-morpholin-4-yl-propan-2-ol, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.826 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6FDC
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-32a | | Descriptor: | (2~{R})-1-[3-[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]pyrazol-1-yl]-3-morpholin-4-yl-propan-2-ol, (2~{S})-1-[5-[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]pyrazol-1-yl]-3-morpholin-4-yl-propan-2-ol, 1,2-ETHANEDIOL, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
8EXX
 
 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and foscarnet (pre-translocation state) | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2022-10-26 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
7TDZ
 
 | |
8HPT
 
 | | Structure of C5a-pep bound mouse C5aR1 in complex with Go | | Descriptor: | Antibody fragment ScFv16, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Saha, S, Maharana, J, Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
6FTX
 
 | | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromatin-remodeling ATPase, ... | | Authors: | Sundaramoorthy, R, Owen-hughes, T, Norman, D.G, Hughes, A. | | Deposit date: | 2018-02-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Elife, 7, 2018
|
|
6SGX
 
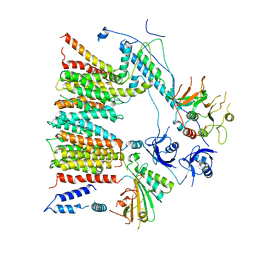 | | Structure of protomer 1 of the ESX-3 core complex | | Descriptor: | ESX-3 secretion system EccB3, ESX-3 secretion system protein EccC3, ESX-3 secretion system protein EccD3, ... | | Authors: | Famelis, N, Rivera-Calzada, A, Llorca, O, Geibel, S. | | Deposit date: | 2019-08-05 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of the mycobacterial type VII secretion system.
Nature, 576, 2019
|
|
5HNY
 
 | | Structural basis of backwards motion in kinesin-14: plus-end directed nKn669 in the AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
6SGY
 
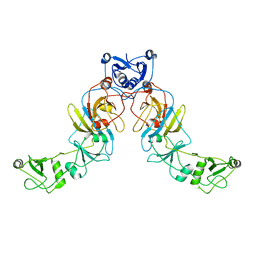 | |
7MPA
 
 | |
6FM6
 
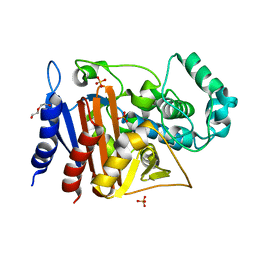 | | Crystal structure of the class C beta-lactamase TRU-1 from Aeromonas enteropelogenes | | Descriptor: | Beta-lactamase, SULFATE ION, TRIETHYLENE GLYCOL | | Authors: | Pozzi, C, De Luca, F, Di Pisa, F, Benvenuti, M, Docquier, J.D, Mangani, S. | | Deposit date: | 2018-01-30 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic-Resolution Structure of a Class C beta-Lactamase and Its Complex with Avibactam.
ChemMedChem, 13, 2018
|
|
6SGW
 
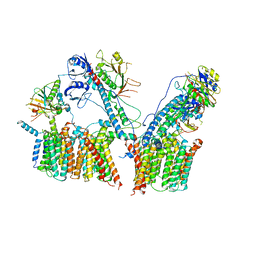 | | Structure of the ESX-3 core complex | | Descriptor: | ESX-3 secretion system ATPase EccB3, ESX-3 secretion system protein EccC3, ESX-3 secretion system protein EccD3, ... | | Authors: | Famelis, N, Rivera-Calzada, A, Llorca, O, Geibel, S. | | Deposit date: | 2019-08-05 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Architecture of the mycobacterial type VII secretion system.
Nature, 576, 2019
|
|
6FM7
 
 | | Crystal structure of the class C beta-lactamase TRU-1 from Aeromonas enteropelogenes in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Pozzi, C, De Luca, F, Benvenuti, M, Di Pisa, F, Docquier, J.D, Mangani, S. | | Deposit date: | 2018-01-30 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Atomic-Resolution Structure of a Class C beta-Lactamase and Its Complex with Avibactam.
ChemMedChem, 13, 2018
|
|
6SGZ
 
 | | Structure of protomer 2 of the ESX-3 core complex | | Descriptor: | ESX-3 secretion system ATPase EccB3, ESX-3 secretion system protein EccC3, ESX-3 secretion system protein EccD3, ... | | Authors: | Famelis, N, Rivera-Calzada, A, Llorca, O, Geibel, S. | | Deposit date: | 2019-08-05 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the mycobacterial type VII secretion system.
Nature, 576, 2019
|
|
5FJA
 
 | | Cryo-EM structure of yeast RNA polymerase III at 4.7 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
6VPO
 
 | | Cryo-EM structure of microtubule-bound KLP61F motor domain in the AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein Klp61F, ... | | Authors: | Bodrug, T, Wilson-Kubalek, E.M, Nithianantham, S, Debs, G, Sindelar, C.V, Milligan, R, Al-Bassam, J. | | Deposit date: | 2020-02-04 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The kinesin-5 tail domain directly modulates the mechanochemical cycle of the motor domain for anti-parallel microtubule sliding.
Elife, 9, 2020
|
|
2UZK
 
 | | Crystal structure of the human FOXO3a-DBD bound to DNA | | Descriptor: | 5'-D(*CP*TP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*AP*G)-3', FORKHEAD BOX PROTEIN O3A | | Authors: | Tsai, K.-L, Sun, Y.-J, Huang, C.-Y, Yang, J.-Y, Hung, M.-C, Hsiao, C.-D. | | Deposit date: | 2007-04-30 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Human Foxo3A-Dbd/DNA Complex Suggests the Effects of Post-Translational Modification.
Nucleic Acids Res., 35, 2007
|
|
5FJ9
 
 | | Cryo-EM structure of yeast apo RNA polymerase III at 4.6 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
