6ZUE
 
 | |
3HV3
 
 | | Human p38 MAP Kinase in Complex with RL49 | | Descriptor: | 1-{4-[(6-aminoquinolin-4-yl)amino]phenyl}-3-[3-tert-butyl-1-(3-methylphenyl)-1H-pyrazol-5-yl]urea, GLYCEROL, Mitogen-activated protein kinase 14, ... | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2009-06-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Displacement assay for the detection of stabilizers of inactive kinase conformations.
J.Med.Chem., 53, 2010
|
|
6ZV5
 
 | | CML1 crystal structure in complex with Lewis a tetrasaccharide | | Descriptor: | Mucin-binding lectin 1, SULFATE ION, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Varrot, A, Bleuler-Martinez, S. | | Deposit date: | 2020-07-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-function relationship of a novel fucoside-binding fruiting body lectin from Coprinopsis cinerea exhibiting nematotoxic activity.
Glycobiology, 32, 2022
|
|
3HUC
 
 | | Human p38 MAP Kinase in Complex with RL40 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase 14, N-[2-phenyl-4-(1H-pyrazol-3-ylamino)quinazolin-7-yl]prop-2-enamide, ... | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2009-06-13 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fluorophore labeling of the glycine-rich loop as a method of identifying inhibitors that bind to active and inactive kinase conformations.
J.Am.Chem.Soc., 132, 2010
|
|
3HV7
 
 | | Human p38 MAP Kinase in Complex with RL38 | | Descriptor: | 1-[1-(3-aminophenyl)-3-tert-butyl-1H-pyrazol-5-yl]-3-naphthalen-1-ylurea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2009-06-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Displacement assay for the detection of stabilizers of inactive kinase conformations.
J.Med.Chem., 53, 2010
|
|
6EPC
 
 | | Ground state 26S proteasome (GS2) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
1WBV
 
 | | Identification of novel p38 alpha MAP Kinase inhibitors using fragment-based lead generation. | | Descriptor: | 3-FLUORO-N-1H-INDOL-5-YL-5-MORPHOLIN-4-YLBENZAMIDE, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tickle, J, Cleasby, A, Devine, L.A, Jhoti, H. | | Deposit date: | 2004-11-05 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation.
J.Med.Chem., 48, 2005
|
|
1WBP
 
 | | SRPK1 bound to 9mer docking motif peptide | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, MEMBRANE-ASSOCIATED GUANYLATE KINASE, ... | | Authors: | Ngo, J.C, Gullinsgrud, J, Chakrabarti, S, Nolen, B, Aubol, B.E, Fu, X.-D, Adams, J.A, McCammon, J.A, Ghosh, G. | | Deposit date: | 2004-11-04 | | Release date: | 2005-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interplay between Srpk and Clk/Sty Kinases in Phosphorylation of the Splicing Factor Asf/Sf2 is Regulated by a Docking Motif in Asf/Sf2
Mol.Cell, 20, 2005
|
|
1WM0
 
 | | PPARgamma in complex with a 2-BABA compound | | Descriptor: | 14-mer from Nuclear receptor coactivator 2, 2-[(2,4-DICHLOROBENZOYL)AMINO]-5-(PYRIMIDIN-2-YLOXY)BENZOIC ACID, Peroxisome proliferator activated receptor gamma | | Authors: | Ostberg, T, Svensson, S, Selen, G, Uppenberg, J, Thor, M, Sundbom, M, Sydow-Backman, M, Gustavsson, A.L, Jendeberg, L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A new class of peroxisome proliferator-activated receptor agonists with a novel binding epitope shows antidiabetic effects
J.Biol.Chem., 279, 2004
|
|
6LQV
 
 | |
6ZV3
 
 | | TFIIS N-terminal domain (TND) from human MED26 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 26 | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZV1
 
 | | TFIIS N-terminal domain (TND) from human IWS1 | | Descriptor: | Protein IWS1 homolog | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZV2
 
 | | TFIIS N-terminal domain (TND) from human PPP1R10 | | Descriptor: | Serine/threonine-protein phosphatase 1 regulatory subunit 10 | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
3IW6
 
 | | Human p38 MAP Kinase in Complex with a Benzylpiperazin-Pyrrol | | Descriptor: | Mitogen-activated protein kinase 14, ethyl 4-[(4-benzylpiperazin-1-yl)carbonyl]-1-ethyl-3,5-dimethyl-1H-pyrrole-2-carboxylate, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
6ZUZ
 
 | |
6ZV4
 
 | | Human TFIIS N-terminal domain in complex with IWS1 | | Descriptor: | Transcription elongation factor A protein 1,Protein IWS1 homolog | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZUY
 
 | | Human TFIIS N-terminal domain (TND) | | Descriptor: | Transcription elongation factor A protein 1 | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
3IHW
 
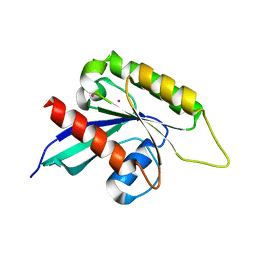 | | Crystal structure of the Ras-like domain of CENTG3 | | Descriptor: | CENTG3, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Tempel, W, Tong, Y, Li, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of the Ras-like domain of CENTG3
to be published
|
|
6LQT
 
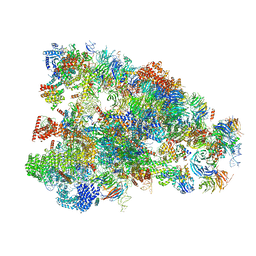 | |
6ERG
 
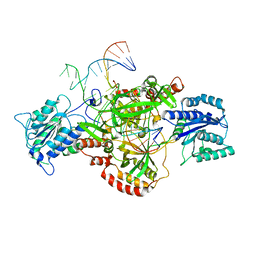 | | Complex of XLF and heterodimer Ku bound to DNA | | Descriptor: | DNA (21-MER), DNA (34-MER), Non-homologous end-joining factor 1, ... | | Authors: | Nemoz, C, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2017-10-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3IP5
 
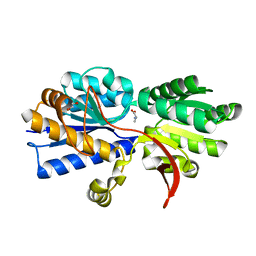 | | Structure of Atu2422-GABA receptor in complex with alanine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), ALANINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IP7
 
 | | Structure of Atu2422-GABA receptor in complex with valine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), CALCIUM ION, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
6LQQ
 
 | |
3IP6
 
 | | Structure of Atu2422-GABA receptor in complex with proline | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), PROLINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IPC
 
 | | Structure of ATU2422-GABA F77A mutant receptor in complex with leucine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), LEUCINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
