3JBM
 
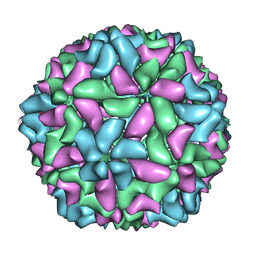 | | Electron cryo-microscopy of a virus-like particle of orange-spotted grouper nervous necrosis virus | | Descriptor: | virus-like particle of orange-spotted grouper nervous necrosis virus | | Authors: | Xie, J, Li, K, Gao, Y, Huang, R, Lai, Y, Shi, Y, Yang, S, Zhu, G, Zhang, Q, He, J. | | Deposit date: | 2015-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural analysis and insertion study reveal the ideal sites for surface displaying foreign peptides on a betanodavirus-like particle
Vet. Res., 47, 2016
|
|
5Z6Y
 
 | | Structure of sfYFP48S95C66BPA | | Descriptor: | Green fluorescent protein | | Authors: | Wang, J.Y, Wang, J.Y. | | Deposit date: | 2018-01-25 | | Release date: | 2019-06-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | structure of sfYFP48S95C66BPA at 1.95 Angstroms resolution
To Be Published
|
|
4M48
 
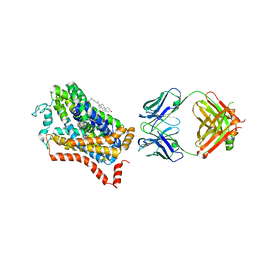 | | X-ray structure of dopamine transporter elucidates antidepressant mechanism | | Descriptor: | 9D5 antibody, heavy chain, light chain, ... | | Authors: | Gouaux, E, Penmatsa, A, Wang, K. | | Deposit date: | 2013-08-06 | | Release date: | 2013-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | X-ray structure of dopamine transporter elucidates antidepressant mechanism.
Nature, 503, 2013
|
|
8H0L
 
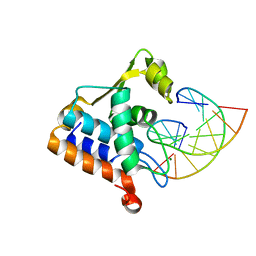 | | Sulfur binding domain of Hga complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*GP*(PST)P*TP*CP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*GP*AP*AP*CP*TP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Liu, G, He, X, Hu, W, Yang, B, Xiao, Q. | | Deposit date: | 2022-09-29 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a promiscuous DNA sulfur binding domain and application in site-directed RNA base editing.
Nucleic Acids Res., 51, 2023
|
|
8HAS
 
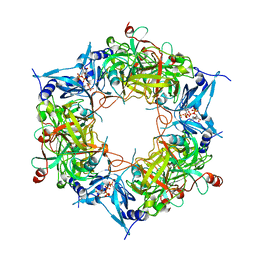 | | NARROW LEAF 1-close from Japonica | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein NARROW LEAF 1 | | Authors: | Zhang, S.J, He, Y.J, Wang, N, Zhang, W.J, Liu, C.M. | | Deposit date: | 2022-10-26 | | Release date: | 2024-05-01 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | NARROW LEAF 1-close from Japonica
To Be Published
|
|
8HAT
 
 | |
4P0M
 
 | | Crystal structure of an evolved putative penicillin-binding protein homolog, Rv2911, from Mycobacterium tuberculosis | | Descriptor: | D-alanyl-D-alanine carboxypeptidase | | Authors: | Krieger, I, Yu, M, Bursey, E, Hung, L.-W, Terwilliger, T.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subfamily-Specific Adaptations in the Structures of Two Penicillin-Binding Proteins from Mycobacterium tuberculosis.
Plos One, 9, 2014
|
|
7BWN
 
 | | Crystal Structure of a Designed Protein Heterocatenane | | Descriptor: | Cellular tumor antigen p53, Chimera of Green fluorescent protein and p53dim | | Authors: | Liu, Y.J, Duan, Z.L, Fang, J, Zhang, F, Xiao, J.Y, Zhang, W.B. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Cellular Synthesis and X-ray Crystal Structure of a Designed Protein Heterocatenane.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5XUY
 
 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13 | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
5XV3
 
 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13, DI(HYDROXYETHYL)ETHER | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
5XV6
 
 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13 | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
5XV4
 
 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13 | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
4XBR
 
 | | In cellulo Crystal Structure of PAK4 in complex with Inka | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein FAM212A,Serine/threonine-protein kinase PAK 4 | | Authors: | Baskaran, Y, Ang, K.C, Anekal, P.V, Chan, W.L, Grimes, J.M, Manser, E, Robinson, R.C. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1
Nat Commun, 6, 2015
|
|
4WFC
 
 | | Structure of the Rrp6-Rrp47 interaction | | Descriptor: | Exosome complex exonuclease RRP6, Exosome complex protein LRP1, SULFATE ION | | Authors: | Schuch, B, Conti, E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase.
Embo J., 33, 2014
|
|
4XBU
 
 | | In vitro Crystal Structure of PAK4 in complex with Inka peptide | | Descriptor: | Protein FAM212A, Serine/threonine-protein kinase PAK 4 | | Authors: | Baskaran, Y, Ang, K.C, Anekal, P.V, Chan, W.L, Grimes, J.M, Manser, E, Robinson, R.C. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1
Nat Commun, 6, 2015
|
|
6D38
 
 | |
6F2G
 
 | | Bacterial asc transporter crystal structure in open to in conformation | | Descriptor: | Nanobody 74, Putative amino acid/polyamine transport protein, ZINC ION | | Authors: | Fort, J, Errasti-Murugarren, E, Carpena, X, Palacin, M, Fita, I. | | Deposit date: | 2017-11-24 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | L amino acid transporter structure and molecular bases for the asymmetry of substrate interaction.
Nat Commun, 10, 2019
|
|
3QAY
 
 | | Catalytic domain of CD27L endolysin targeting Clostridia Difficile | | Descriptor: | Endolysin, PHOSPHATE ION, ZINC ION | | Authors: | Mayer, M.J, Garefaliki, V, Spoerl, R, Narbad, A, Meijers, R. | | Deposit date: | 2011-01-12 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based modification of a Clostridium difficile-targeting endolysin affects activity and host range.
J.Bacteriol., 193, 2011
|
|
6FP9
 
 | | Crystal structure of anti-mTFP1 DARPin 1238_E11 | | Descriptor: | 1,2-ETHANEDIOL, DARPin 1238_E11, SULFATE ION | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
3RHB
 
 | | Crystal structure of the apo form of glutaredoxin C5 from Arabidopsis thaliana | | Descriptor: | GLUTATHIONE, Glutaredoxin-C5, chloroplastic, ... | | Authors: | Roret, T, Couturier, J, Tsan, P, Jacquot, J.P, Rouhier, N, Didierjean, C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Arabidopsis chloroplastic glutaredoxin c5 as a model to explore molecular determinants for iron-sulfur cluster binding into glutaredoxins.
J.Biol.Chem., 286, 2011
|
|
3RHC
 
 | | Crystal structure of the holo form of glutaredoxin C5 from Arabidopsis thaliana | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLUTATHIONE, Glutaredoxin-C5, ... | | Authors: | Roret, T, Couturier, J, Tsan, P, Jacquot, J.P, Rouhier, N, Didierjean, C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Arabidopsis chloroplastic glutaredoxin c5 as a model to explore molecular determinants for iron-sulfur cluster binding into glutaredoxins.
J.Biol.Chem., 286, 2011
|
|
5O1Q
 
 | | LysF1 sh3b domain structure | | Descriptor: | sh3b domain | | Authors: | Benesik, M, Novacek, J, Janda, L, Dopitova, R, Pernisova, M, Melkova, K, Tisakova, L, Doskar, J, Zidek, L, Hejatko, J, Pantucek, R. | | Deposit date: | 2017-05-19 | | Release date: | 2017-09-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Role of SH3b binding domain in a natural deletion mutant of Kayvirus endolysin LysF1 with a broad range of lytic activity.
Virus Genes, 54, 2018
|
|
7DIG
 
 | |
6NHT
 
 | |
5NHN
 
 | |
