3KV0
 
 | | Crystal structure of HET-C2: A FUNGAL GLYCOLIPID TRANSFER PROTEIN (GLTP) | | Descriptor: | HET-C2 | | Authors: | Simanshu, D.K, Kenoth, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2009-11-28 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determination and tryptophan fluorescence of heterokaryon incompatibility C2 protein (HET-C2), a fungal glycolipid transfer protein (GLTP), provide novel insights into glycolipid specificity and membrane interaction by the GLTP fold.
J.Biol.Chem., 285, 2010
|
|
1P9R
 
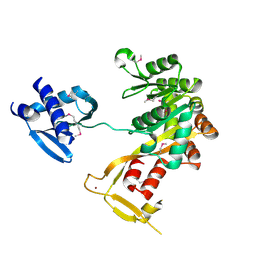 | | Crystal Structure of Vibrio cholerae putative NTPase EpsE | | Descriptor: | CHLORIDE ION, General secretion pathway protein E, ZINC ION | | Authors: | Robien, M.A, Krumm, B.E, Sandkvist, M, Hol, W.G.J. | | Deposit date: | 2003-05-12 | | Release date: | 2003-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae
J.Mol.Biol., 333, 2003
|
|
3KQL
 
 | |
1N4D
 
 | |
1UPV
 
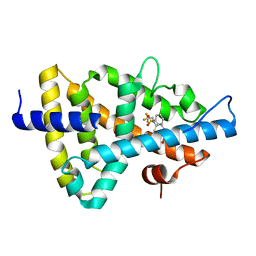 | | Crystal structure of the human Liver X receptor beta ligand binding domain in complex with a synthetic agonist | | Descriptor: | N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, OXYSTEROLS RECEPTOR LXR-BETA | | Authors: | Hoerer, S, Schmid, A, Heckel, A, Budzinski, R.M, Nar, H. | | Deposit date: | 2003-10-13 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human Liver X Receptor Beta Ligand-Binding Domain in Complex with a Synthetic Agonist
J.Mol.Biol., 334, 2003
|
|
4BX0
 
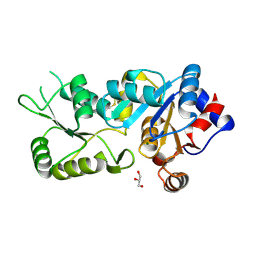 | | Crystal Structure of a Monomeric Variant of murine Chronophin (Pyridoxal Phosphate phosphatase) | | Descriptor: | GLYCEROL, MAGNESIUM ION, PYRIDOXAL PHOSPHATE PHOSPHATASE | | Authors: | Kestler, C, Knobloch, G, Gohla, A, Schindelin, H. | | Deposit date: | 2013-07-08 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Chronophin Dimerization is Required for Proper Positioning of its Substrate Specificity Loop
J.Biol.Chem., 289, 2014
|
|
3Q3Q
 
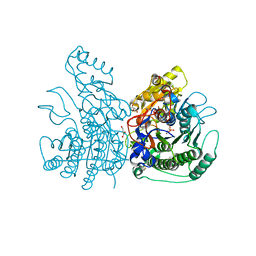 | |
3LJD
 
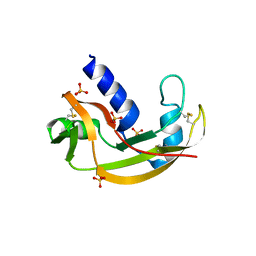 | | The X-ray structure of zebrafish RNase1 from a new crystal form at pH 4.5 | | Descriptor: | ACETATE ION, SULFATE ION, Zebrafish RNase1 | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-12-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish.
Biochem.J., 433, 2010
|
|
5ZBH
 
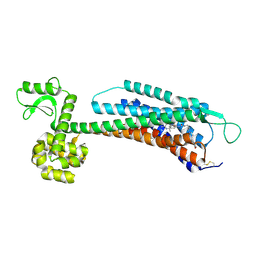 | | The Crystal Structure of Human Neuropeptide Y Y1 Receptor with BMS-193885 | | Descriptor: | Neuropeptide Y receptor type 1,T4 Lysozyme,Neuropeptide Y receptor type 1, dimethyl 4-{3-[({3-[4-(3-methoxyphenyl)piperidin-1-yl]propyl}carbamoyl)amino]phenyl}-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate | | Authors: | Yang, Z, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2018-02-11 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of ligand binding modes at the neuropeptide Y Y1receptor
Nature, 556, 2018
|
|
1NF2
 
 | |
4IFW
 
 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, ADP inhibited form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Thiamine biosynthesis lipoprotein ApbE | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3001 Å) | | Cite: | The TP0796 Lipoprotein of Treponema pallidum Is a Bimetal-dependent FAD Pyrophosphatase with a Potential Role in Flavin Homeostasis.
J.Biol.Chem., 288, 2013
|
|
4IG1
 
 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, Mg(II)-AMP product bound form | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, FAD:protein FMN transferase, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4318 Å) | | Cite: | The TP0796 Lipoprotein of Treponema pallidum Is a Bimetal-dependent FAD Pyrophosphatase with a Potential Role in Flavin Homeostasis.
J.Biol.Chem., 288, 2013
|
|
1OJL
 
 | |
3LQY
 
 | | Crystal structure of putative isochorismatase hydrolase from Oleispira antarctica | | Descriptor: | GLYCEROL, putative isochorismatase hydrolase | | Authors: | Goral, A, Chruszcz, M, Kagan, O, Cymborowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-10 | | Release date: | 2010-03-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative isochorismatase hydrolase from Oleispira antarctica.
J.Struct.Funct.Genom., 13, 2012
|
|
4IFX
 
 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, FAD substrate bound form | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | The TP0796 Lipoprotein of Treponema pallidum Is a Bimetal-dependent FAD Pyrophosphatase with a Potential Role in Flavin Homeostasis.
J.Biol.Chem., 288, 2013
|
|
5ZBQ
 
 | | The Crystal Structure of human neuropeptide Y Y1 receptor with UR-MK299 | | Descriptor: | Neuropeptide Y receptor type 1,T4 Lysozyme, N~2~-(diphenylacetyl)-N-[(4-hydroxyphenyl)methyl]-N~5~-(N'-{[2-(propanoylamino)ethyl]carbamoyl}carbamimidoyl)-D-ornithinamide | | Authors: | Yang, Z, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2018-02-12 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand binding modes at the neuropeptide Y Y1receptor
Nature, 556, 2018
|
|
3LYK
 
 | |
4JEM
 
 | | Crystal structure of MilB complexed with cytidine 5'-monophosphate | | Descriptor: | CMP/hydroxymethyl CMP hydrolase, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Sikowitz, M.D, Cooper, L.E, Begley, T.P, Kaminski, P.A, Ealick, S.E. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Reversal of the substrate specificity of CMP N-glycosidase to dCMP.
Biochemistry, 52, 2013
|
|
2CLB
 
 | | The structure of the DPS-like protein from Sulfolobus solfataricus reveals a bacterioferritin-like di-metal binding site within a Dps- like dodecameric assembly | | Descriptor: | DPS-LIKE PROTEIN, FE (III) ION, ZINC ION | | Authors: | Gauss, G.H, Benas, P, Wiedenheft, B, Young, M, Douglas, T, Lawrence, C.M. | | Deposit date: | 2006-04-26 | | Release date: | 2006-07-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Dps-Like Protein from Sulfolobus Solfataricus Reveals a Bacterioferritin-Like Dimetal Binding Site within a Dps-Like Dodecameric Assembly.
Biochemistry, 45, 2006
|
|
1RLF
 
 | | STRUCTURE DETERMINATION OF THE RAS-BINDING DOMAIN OF THE RAL-SPECIFIC GUANINE NUCLEOTIDE EXCHANGE FACTOR RLF, NMR, 10 STRUCTURES | | Descriptor: | RLF | | Authors: | Esser, D, Bauer, B, Wolthuis, R.M.F, Wittinghofer, A, Cool, R.H, Bay, P. | | Deposit date: | 1998-07-09 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure determination of the Ras-binding domain of the Ral-specific guanine nucleotide exchange factor Rlf.
Biochemistry, 37, 1998
|
|
1S7F
 
 | | RimL- Ribosomal L7/L12 alpha-N-protein acetyltransferase crystal form I (apo) | | Descriptor: | CHLORIDE ION, MALONIC ACID, acetyl transferase | | Authors: | Vetting, M.W, de Carvalho, L.P, Roderick, S.L, Blanchard, J.S. | | Deposit date: | 2004-01-29 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel dimeric structure of the RimL Nalpha-acetyltransferase from Salmonella typhimurium.
J.Biol.Chem., 280, 2005
|
|
9AX8
 
 | | 70S initiation complex (tRNA-fMet M1, initiation factor 2 + CUG start codon) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mattingly, J.M, Nguyen, H.A, Dunham, C.M. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural analysis of noncanonical translation initiation complexes.
J.Biol.Chem., 2024
|
|
9EQF
 
 | | Crystal structure of the L-arginine hydroxylase VioC MeHis316, bound to Fe(II), L-arginine, and succinate | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | Authors: | Hardy, F.J. | | Deposit date: | 2024-03-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing Ferryl Reactivity in a Nonheme Iron Oxygenase Using an Expanded Genetic Code.
Acs Catalysis, 14, 2024
|
|
8X77
 
 | | Enterovirus proteinase with host factor | | Descriptor: | 2A protein, Actin-histidine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2023-11-23 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | The EV71 2A protease occupies the central cleft of SETD3 and disrupts SETD3-actin interaction.
Nat Commun, 15, 2024
|
|
9BD2
 
 | | MAGE-A3 MHD crystal soaked with KL861 | | Descriptor: | (furan-2-yl)[4-({(5P)-5-(1H-indazol-4-yl)-2-[3-(morpholin-4-yl)propoxy]phenyl}methyl)piperazin-1-yl]methanone, Melanoma-associated antigen 3 | | Authors: | Butrin, A, Krone, M.W, Li, K, Bond, M.J, Linhares, B.M, Crews, C. | | Deposit date: | 2024-04-10 | | Release date: | 2024-09-18 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Development of Ligands and Degraders Targeting MAGE-A3.
J.Am.Chem.Soc., 146, 2024
|
|
