6LOG
 
 | |
6EXT
 
 | |
7ALN
 
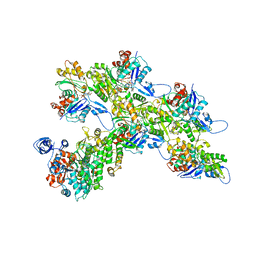 | | Cryo-EM structure of the divergent actomyosin complex from Plasmodium falciparum Myosin A in the Rigor state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-1, Jasplakinolide, ... | | Authors: | Robert-Paganin, J, Xu, X.-P, Swift, M.F, Auguin, D, Robblee, J.P, Lu, H, Fagnant, P.M, Krementsova, E.B, Trybus, K.M, Houdusse, A, Volkmann, N, Hanein, D. | | Deposit date: | 2020-10-06 | | Release date: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | The actomyosin interface contains an evolutionary conserved core and an ancillary interface involved in specificity.
Nat Commun, 12, 2021
|
|
6EXU
 
 | |
7QG9
 
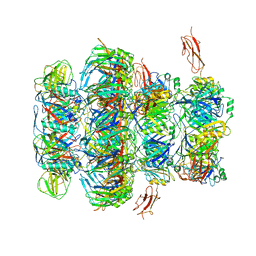 | | Tail tip of siphophage T5 : common core proteins | | Descriptor: | Distal tail protein, L-shaped tail fiber protein p132, Minor tail protein, ... | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-21 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
3GVM
 
 | |
4FP4
 
 | | Crystal structure of isoprenoid synthase a3mx09 (target efi-501993) from pyrobaculum calidifontis | | Descriptor: | GERAN-8-YL GERAN, Polyprenyl synthetase, UNKNOWN ATOM OR ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-06-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F62
 
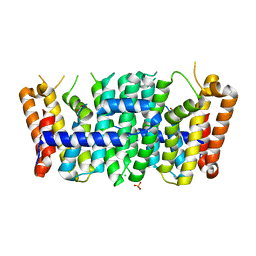 | | Crystal structure of a putative farnesyl-diphosphate synthase from Marinomonas sp. MED121 (Target EFI-501980) | | Descriptor: | CHLORIDE ION, GLYCEROL, Geranyltranstransferase, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1VHP
 
 | | VH-P8, NMR | | Descriptor: | VH-P8 | | Authors: | Riechmann, L. | | Deposit date: | 1996-04-16 | | Release date: | 1996-11-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Rearrangement of the former VL interface in the solution structure of a camelised, single antibody VH domain.
J.Mol.Biol., 259, 1996
|
|
5VSI
 
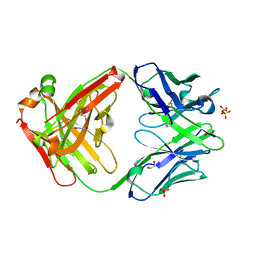 | | CH1/Ckappa Fab mutant 15.1 | | Descriptor: | 1,2-ETHANEDIOL, CH1/Ckappa Fab heavy chain, CH1/Ckappa Fab light chain, ... | | Authors: | Hendle, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Computational design of a specific heavy chain/ kappa light chain interface for expressing fully IgG bispecific antibodies.
Protein Sci., 26, 2017
|
|
7RTY
 
 | |
3HK5
 
 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Arabinarate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-arabinaric acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-05-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3HK8
 
 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Arabinohydroxamate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-arabinohydroxamic acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-05-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3HKA
 
 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Fructuronate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-fructuronic acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-05-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
6O47
 
 | | human cGAS core domain (K427E/K428E) bound with RU-521 | | Descriptor: | (3~{S})-3-[1-[4,5-bis(chloranyl)-1~{H}-benzimidazol-2-yl]-3-methyl-5-oxidanyl-pyrazol-4-yl]-3~{H}-2-benzofuran-1-one, 2-(4,5-dichloro-1H-benzimidazol-2-yl)-5-methyl-4-[(1R)-3-oxo-1,3-dihydro-2-benzofuran-1-yl]-1,2-dihydro-3H-pyrazol-3-one, CITRIC ACID, ... | | Authors: | Xie, W, Lama, L, Adura, C, Glickman, J.F, Tuschl, T, Patel, D.J. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3WOH
 
 | |
5FNZ
 
 | |
5FO1
 
 | |
5FO0
 
 | |
1RDF
 
 | | G50P mutant of phosphonoacetaldehyde hydrolase in complex with substrate analogue vinyl sulfonate | | Descriptor: | ETHANESULFONIC ACID, MAGNESIUM ION, phosphonoacetaldehyde hydrolase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-11-05 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the substrate specificity loop of the HAD superfamily cap domain
Biochemistry, 43, 2004
|
|
5DC0
 
 | |
5DC9
 
 | | CRYSTAL STRUCTURE OF MONOBODY AS25/ABL1 SH2 DOMAIN COMPLEX, CRYSTAL B | | Descriptor: | AS25 monobody, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wojcik, J.B, Koide, A, Koide, S. | | Deposit date: | 2015-08-23 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Allosteric Inhibition of Bcr-Abl Kinase by High Affinity Monobody Inhibitors Directed to the Src Homology 2 (SH2)-Kinase Interface.
J.Biol.Chem., 291, 2016
|
|
4X0R
 
 | | Crystal structure of human MxB stalk domain | | Descriptor: | Interferon-induced GTP-binding protein Mx2 | | Authors: | Yu, X.-F, Xie, W. | | Deposit date: | 2014-11-23 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | Structural insight into the assembly of human anti-HIV dynamin-like protein MxB/Mx2.
Biochem.Biophys.Res.Commun., 456, 2015
|
|
6VIE
 
 | | Structure of caspase-1 in complex with gasdermin D | | Descriptor: | Caspase-1 subunit p10, Caspase-1 subunit p20, Gasdermin-D | | Authors: | Liu, Z, Xiao, T.S. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Caspase-1 Engages Full-Length Gasdermin D through Two Distinct Interfaces That Mediate Caspase Recruitment and Substrate Cleavage.
Immunity, 53, 2020
|
|
5KF6
 
 | |
