4UA7
 
 | |
8EQZ
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog T0-C6 | | Descriptor: | N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-N-hexylbenzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Li, Y, Chen, T. | | Deposit date: | 2022-10-11 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8E3N
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with rifamycin S | | Descriptor: | Nuclear receptor subfamily 1 group I member 2, Rifamycin S | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Chen, T. | | Deposit date: | 2022-08-17 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FPE
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog T0-BP | | Descriptor: | N-[([1,1'-biphenyl]-4-yl)methyl]-N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Li, Y, Chen, T. | | Deposit date: | 2023-01-04 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7UVL
 
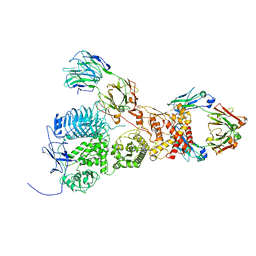 | | IgA1 Protease with IgA1 substrate | | Descriptor: | Immunoglobulin alpha-1 heavy chain, Immunoglobulin alpha-1 heavy constant, Immunoglobulin alpha-1 light chain, ... | | Authors: | Eisenmesser, Z.E, Zheng, H. | | Deposit date: | 2022-05-02 | | Release date: | 2022-11-23 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | A substrate-induced gating mechanism is conserved among Gram-positive IgA1 metalloproteases.
Commun Biol, 5, 2022
|
|
4UA9
 
 | |
4UAA
 
 | |
4UA6
 
 | |
3K82
 
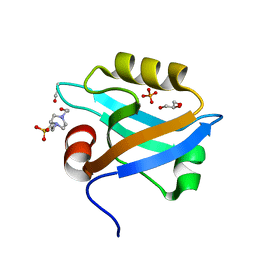 | | Crystal Structure of the third PDZ domain of PSD-95 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Disks large homolog 4, GLYCEROL, ... | | Authors: | Camara-Artigas, A, Gavira, J.A. | | Deposit date: | 2009-10-13 | | Release date: | 2010-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel conformational aspects of the third PDZ domain of the neuronal post-synaptic density-95 protein revealed from two 1.4A X-ray structures
J.Struct.Biol., 170, 2010
|
|
8ATG
 
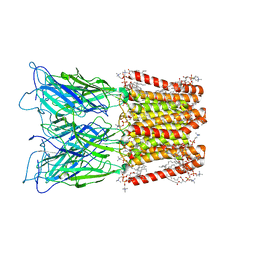 | | Pentameric ligand-gated ion channel GLIC with bound lipids | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Proton-gated ion channel | | Authors: | Bergh, C, Rovsnik, U, Howard, R.J, Lindahl, E. | | Deposit date: | 2022-08-23 | | Release date: | 2023-09-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery of lipid binding sites in a ligand-gated ion channel by integrating simulations and cryo-EM.
Elife, 12, 2024
|
|
3CLD
 
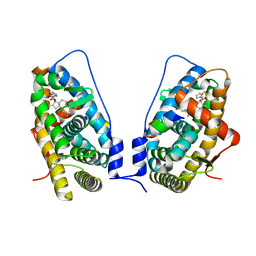 | | Ligand binding domain of the glucocorticoid receptor complexed with fluticazone furoate | | Descriptor: | (6alpha,11alpha,14beta,16alpha,17alpha)-6,9-difluoro-17-{[(fluoromethyl)sulfanyl]carbonyl}-11-hydroxy-16-methyl-3-oxoan drosta-1,4-dien-17-yl furan-2-carboxylate, Glucocorticoid receptor, Tif2 coactivator motif | | Authors: | Shewchuk, L.M, McLay, I, Stewart, E, Biggadike, K.B, Hassell, A.M, Bledsoe, R.K. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | X-ray crystal structure of the novel enhanced-affinity glucocorticoid agonist fluticasone furoate in the glucocorticoid receptor-ligand binding domain.
J.Med.Chem., 51, 2008
|
|
5J9I
 
 | |
5Q1B
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-{[(2S)-2-cyclohexyl-2-{5,6-difluoro-2-[4-(1,3-thiazol-2-yl)phenyl]-1H-benzimidazol-1-yl}acetyl]amino}benzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0I
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 2-[(3,4-dimethoxyphenyl)-(4-methylphenyl)sulfonyl-amino]-N-(2,4-dimethylpentan-3-yl)ethanamide, Bile acid receptor, COACTIVATOR PEPTIDE PGC-1A PPAR GAMMA COACTIVATOR | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0Q
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, ethyl 4-({2-phenyl-5-[(thiophen-2-yl)sulfonyl]-4,5,6,7-tetrahydro-2H-pyrazolo[4,3-c]pyridine-3-carbonyl}amino)benzoate | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q13
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-[6-chloro-2-(4-chlorophenyl)-5-fluoro-1H-benzimidazol-1-yl]-N-cyclohexyl-2-[(2S)-oxan-2-yl]acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0M
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 5-{[(3beta,5beta,14beta,17alpha)-3-hydroxy-24-oxocholan-24-yl]amino}benzene-1,3-dicarboxylic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0W
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-({5-bromo-1'-[(2-chlorophenyl)sulfonyl]-2-oxospiro[indole-3,4'-piperidin]-1(2H)-yl}methyl)benzoic acid, Bile acid receptor, cDNA FLJ76652, ... | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q12
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 2-(2,6-difluorophenyl)-N-(2,6-dimethylphenyl)-5-methylimidazo[1,2-a]pyridin-3-amine, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1G
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2E)-N-cyclohexyl-N-(cyclohexylcarbamoyl)-3-(4-fluorophenyl)prop-2-enamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0U
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, trans-4-({(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylacetyl}amino)cyclohexyl hydrogen sulfate | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1C
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-cyclohexyl-2-[2-(2,6-dimethoxypyridin-3-yl)-5,6-difluoro-1H-benzimidazol-1-yl]-N-(trans-4-hydroxycyclohexyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0O
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-{2-[4-(benzenecarbonyl)phenyl]-1H-benzimidazol-1-yl}-N,2-dicyclohexylacetamide, Bile acid receptor, CHLORIDE ION, ... | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q14
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-{(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylethoxy}-3-fluorobenzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
8S4J
 
 | | Structure, substrate selectivity determinants and membrane interactions of a Glutamate-specific TAXI TRAP binding protein from Vibrio cholerae. | | Descriptor: | GLUTAMIC ACID, SODIUM ION, TAXI family TRAP transporter solute-binding subunit | | Authors: | Mulligan, C, Daab, A, Davies, J. | | Deposit date: | 2024-02-21 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and selectivity of a glutamate-specific TAXI TRAP binding protein from Vibrio cholerae.
J.Gen.Physiol., 156, 2024
|
|
