2XA6
 
 | | Structural basis for homodimerization of the Src-associated during mitosis, 68 kD protein (Sam68) Qua1 domain | | Descriptor: | KH DOMAIN-CONTAINING,RNA-BINDING,SIGNAL TRANSDUCTION-ASSOCIATED PROTEIN 1 | | Authors: | Meyer, N.H, Tripsianes, K, Vincendeaux, M, Madl, T, Kateb, F, Brack-Werner, R, Sattler, M. | | Deposit date: | 2010-03-29 | | Release date: | 2010-07-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for homodimerization of the Src-associated during mitosis, 68-kDa protein (Sam68) Qua1 domain.
J. Biol. Chem., 285, 2010
|
|
2X43
 
 | | STRUCTURAL BASIS OF MOLECULAR RECOGNITION BY SHERP AT MEMBRANE SURFACES | | Descriptor: | SHERP | | Authors: | Moore, B, Miles, A.J, Guerra, C.G, Simpson, P, Iwata, M, Wallace, B.A, Matthews, S.J, Smith, D.F, Brown, K.A. | | Deposit date: | 2010-02-09 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Moelcular Recognition by the Leishmania Small Hydrophilic Endoplasmic Reticulum-Associated Protein, Sherp, at Membrane Surfaces
J.Biol.Chem., 286, 2011
|
|
6ES5
 
 | |
6DZC
 
 | |
6DZA
 
 | |
6DZE
 
 | |
6EZ0
 
 | | Specific phosphorothioate substitution within domain 6 of a group II intron ribozyme leads to changes in local structure and metal ion binding | | Descriptor: | RNA (27-MER) | | Authors: | Erat, M.C, Besic, E, Oberhuber, M, Johannsen, S, Sigel, R.K.O. | | Deposit date: | 2017-11-13 | | Release date: | 2018-01-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Specific phosphorothioate substitution within domain 6 of a group II intron ribozyme leads to changes in local structure and metal ion binding.
J. Biol. Inorg. Chem., 23, 2018
|
|
6F2X
 
 | | Structural characterization of the Mycobacterium tuberculosis Protein Tyrosine Kinase A (PtkA) | | Descriptor: | Protein Tyrosine Kinase A | | Authors: | Niesteruk, A, Jonker, H.R.A, Sreeramulu, S, Richter, C, Hutchison, M, Linhard, V, Schwalbe, H. | | Deposit date: | 2017-11-27 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The domain architecture of PtkA, the first tyrosine kinase fromMycobacterium tuberculosis, differs from the conventional kinase architecture.
J. Biol. Chem., 293, 2018
|
|
6FC9
 
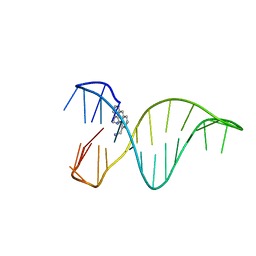 | | The 1,8-bis(aminomethyl)anthracene and Quadruplex-duplex junction complex | | Descriptor: | DNA (27-MER), [8-(azaniumylmethyl)anthracen-1-yl]methylazanium | | Authors: | Santana, A, Serrano, I, Montalvillo-Jimenez, L, Corzana, F, Bastida, A, Jimenez-Barbero, J, Gonzalez, C, Asensio, J.L. | | Deposit date: | 2017-12-20 | | Release date: | 2019-04-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | De Novo Design of Selective Quadruplex-Duplex Junction Ligands and Structural Characterisation of Their Binding Mode: Targeting the G4 Hot-Spot.
Chemistry, 2020
|
|
6DAT
 
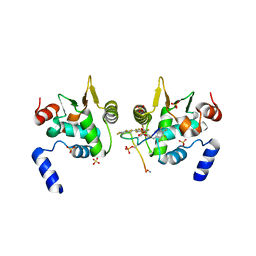 | | ETS1 in complex with synthetic SRR mimic | | Descriptor: | Protein C-ets-1, SULFATE ION, serine-rich region (SRR) peptide | | Authors: | Perez-Borrajero, C, Okon, M, Lin, C.S, Scheu, K, Murphy, M.E.P, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2018-05-02 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35002637 Å) | | Cite: | The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
J. Mol. Biol., 431, 2019
|
|
6FY6
 
 | | Concerted dynamics of metallo-base pairs in an A/B-form helical transition (major species) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*TP*CP*AP*TP*GP*AP*TP*AP*CP*G)-3')_major, MERCURY (II) ION | | Authors: | Schmidt, O.P, Jurt, S, Johannsen, S, Karimi, A, Sigel, R.K.O, Luedtke, N.W. | | Deposit date: | 2018-03-11 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Concerted dynamics of metallo-base pairs in an A/B-form helical transition.
Nat Commun, 10, 2019
|
|
6GDZ
 
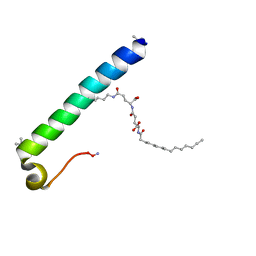 | | exendin-4 based dual GLP-1/glucagon receptor agonist | | Descriptor: | (2~{S})-2-[[(4~{S})-4-(hexadecanoylamino)-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]pentanedioic acid, Exendin-4 | | Authors: | Evers, A, Kurz, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-06-20 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Dual Glucagon-like Peptide 1 (GLP-1)/Glucagon Receptor Agonists Specifically Optimized for Multidose Formulations.
J. Med. Chem., 61, 2018
|
|
6FY7
 
 | | Concerted dynamics of metallo-base pairs in an A/B-form helical transition (minor species) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*TP*CP*AP*TP*GP*AP*TP*AP*CP*G)-3')_minor, MERCURY (II) ION | | Authors: | Schmidt, O.P, Jurt, S, Johannsen, S, Karimi, A, Sigel, R.K.O, Luedtke, N.W. | | Deposit date: | 2018-03-11 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Concerted dynamics of metallo-base pairs in an A/B-form helical transition.
Nat Commun, 10, 2019
|
|
6GE2
 
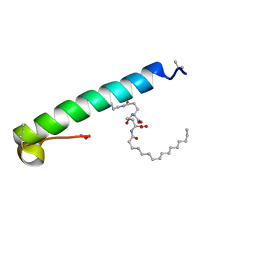 | | exendin-4 based dual GLP-1/glucagon receptor agonist | | Descriptor: | (2~{S})-2-[[(4~{S})-4-(hexadecanoylamino)-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]pentanedioic acid, Exendin-4 | | Authors: | Evers, A, Kurz, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-06-20 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Dual Glucagon-like Peptide 1 (GLP-1)/Glucagon Receptor Agonists Specifically Optimized for Multidose Formulations.
J. Med. Chem., 61, 2018
|
|
6DZB
 
 | |
6GDK
 
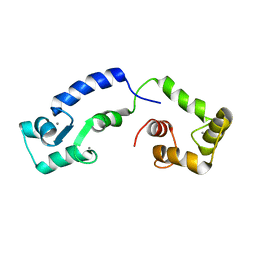 | | Calcium bound form of human calmodulin mutant F141L | | Descriptor: | CALCIUM ION, Calmodulin-1 | | Authors: | Grachov, O, Holt, C, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2018-04-23 | | Release date: | 2018-10-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Arrhythmia mutations in calmodulin cause conformational changes that affect interactions with the cardiac voltage-gated calcium channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2XFM
 
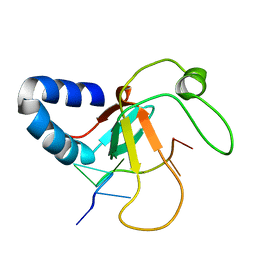 | | Complex structure of the MIWI Paz domain bound to methylated single stranded RNA | | Descriptor: | 5'-R(*AP*CP*CP*GP*AP*CP*UP*(OMU)P)-3', PIWI-LIKE PROTEIN 1 | | Authors: | Simon, B, Kirkpatrick, J.P, Eckhardt, S, Sehr, P, Andrade-Navarro, M.A, Pillai, R.S, Carlomagno, T. | | Deposit date: | 2010-05-27 | | Release date: | 2011-01-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Recognition of 2'-O-Methylated 3'-End of Pirna by the Paz Domain of a Piwi Protein.
Structure, 19, 2011
|
|
1JFP
 
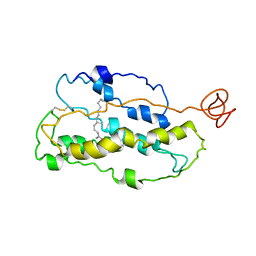 | | Structure of bovine rhodopsin (dark adapted) | | Descriptor: | RETINAL, rhodopsin | | Authors: | Yeagle, P.L, Choi, G, Albert, A.D. | | Deposit date: | 2001-06-21 | | Release date: | 2001-10-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Studies on the structure of the G-protein-coupled receptor rhodopsin including the putative G-protein binding site in unactivated and activated forms.
Biochemistry, 40, 2001
|
|
1YU7
 
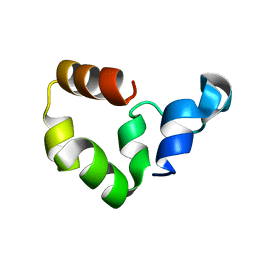 | | Crystal Structure of the W64Y mutant of Villin Headpiece | | Descriptor: | Villin | | Authors: | Meng, J, Vardar, D, Wang, Y, Guo, H.C, Head, J.F, McKnight, C.J. | | Deposit date: | 2005-02-12 | | Release date: | 2005-09-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structures of villin headpiece and mutants with reduced F-actin binding activity.
Biochemistry, 44, 2005
|
|
1LDL
 
 | |
1M23
 
 | |
2AFF
 
 | | The solution structure of the Ki67FHA/hNIFK(226-269)3P complex | | Descriptor: | Antigen KI-67, MKI67 FHA domain interacting nucleolar phosphoprotein | | Authors: | Byeon, I.-J.L, Li, H, Song, H, Gronenborn, A.M, Tsai, M.D. | | Deposit date: | 2005-07-25 | | Release date: | 2005-10-25 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Sequential phosphorylation and multisite interactions characterize specific target recognition by the FHA domain of Ki67.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1XNL
 
 | | ASLV fusion peptide | | Descriptor: | membrane protein gp37 | | Authors: | Cheng, S.F, Wu, C.W, Kantchev, E.A, Chang, D.K. | | Deposit date: | 2004-10-05 | | Release date: | 2005-01-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure and membrane interaction of the internal fusion peptide of avian sarcoma leukosis virus
Eur.J.Biochem., 271, 2004
|
|
1UHC
 
 | | Solution Structure of RSGI RUH-002, a SH3 Domain of KIAA1010 protein [Homo sapiens] | | Descriptor: | KIAA1010 protein | | Authors: | Abe, T, Hirota, H, Kobayashi, N, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-28 | | Release date: | 2003-12-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-002, a SH3 Domain of KIAA1010 protein [Homo sapiens]
To be Published
|
|
1XN7
 
 | | Solution Structure of E.Coli Protein yhgG: The Northeast Structural Genomics Consortium Target ET95 | | Descriptor: | Hypothetical protein yhgG | | Authors: | Shen, Y, Acton, T, Atreya, H.S, Ma, L, Liu, G, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of E.Coli Protein yhgG: The Northeast Structural Genomics Consortium Target ET95
To be Published
|
|
