8CRG
 
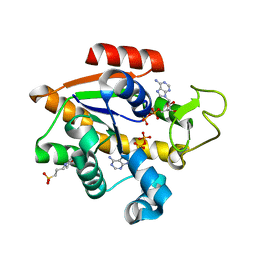 | | E. coli adenylate kinase in complex with two ADP molecules as a result of enzymatic AP4A hydrolysis | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase | | Authors: | Oelker, M, Tischlik, S, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2023-03-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Insights into Enzymatic Catalysis from Binding and Hydrolysis of Diadenosine Tetraphosphate by E. coli Adenylate Kinase.
Biochemistry, 62, 2023
|
|
7PSL
 
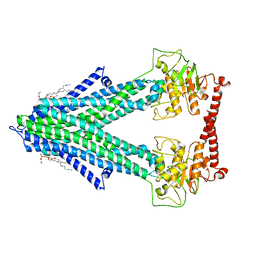 | | S. cerevisiae Atm1 in MSP1D1 nanodiscs in nucleotide-free state | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, Iron-sulfur clusters transporter ATM1, mitochondrial, ... | | Authors: | Ellinghaus, T.L, Kuehlbrandt, W. | | Deposit date: | 2021-09-23 | | Release date: | 2021-12-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational changes in the yeast mitochondrial ABC transporter Atm1 during the transport cycle.
Sci Adv, 7, 2021
|
|
1Q19
 
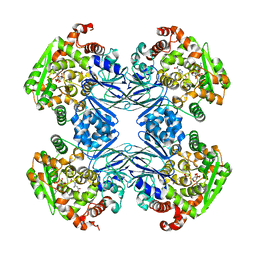 | | Carbapenam Synthetase | | Descriptor: | (2S,5S)-5-CARBOXYMETHYLPROLINE, CarA, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Miller, M.T, Gerratana, B, Stapon, A, Townsend, C.A, Rosenzweig, A.C. | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Carbapenam Synthetase (CarA)
J.Biol.Chem., 278, 2003
|
|
1Q3Q
 
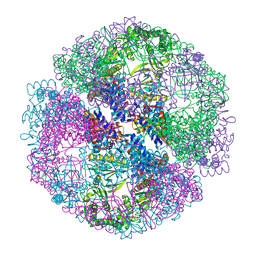 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (two-point mutant complexed with AMP-PNP) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Maruyama, T, Yohda, M, Miki, K. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
3DD7
 
 | |
6MY3
 
 | | Solution structure of gomesin at 310K | | Descriptor: | gomesin | | Authors: | Chin, Y.K.-Y, Deplazes, E. | | Deposit date: | 2018-11-01 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | The unusual conformation of cross-strand disulfide bonds is critical to the stability of beta-hairpin peptides.
Proteins, 88, 2020
|
|
3ROJ
 
 | | D-fructose 1,6-bisphosphatase class 2/sedoheptulose 1,7-bisphosphatase of Synechocystis sp. PCC 6803 | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, D-fructose 1,6-bisphosphatase class 2/sedoheptulose 1,7-bisphosphatase, ... | | Authors: | Hu, X, Hui, D, Lingling, F, Jian, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New insights into the structural and interactional basis for a promising route towards fructose-1,6-/sedoheptulose-1,7-bisphosphatases controlling
To be Published
|
|
5U7X
 
 | | Crystal structure of a nucleoside triphosphate diphosphohydrolase (NTPDase) from the legume Vigna unguiculata subsp. cylindrica (Dolichos biflorus) in complex with phosphate and manganese | | Descriptor: | MANGANESE (II) ION, Nod factor binding lectin-nucleotide phosphohydrolase, PHOSPHATE ION | | Authors: | Cumming, M.H, Summers, E.L, Oulavallickal, T, Roberts, N, Arcus, V.L. | | Deposit date: | 2016-12-12 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures and kinetics for plant nucleoside triphosphate diphosphohydrolases support a domain motion catalytic mechanism.
Protein Sci., 26, 2017
|
|
3EH9
 
 | | Crystal structure of death associated protein kinase complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Death-associated protein kinase 1, SULFATE ION | | Authors: | McNamara, L.K, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2008-09-11 | | Release date: | 2009-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into nucleotide recognition by human death-associated protein kinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3BWF
 
 | | Crystal structure of the human Pim1 in complex with an osmium compound | | Descriptor: | PYRIDOCARBAZOLE CYCLOPENTADIENYL OS(CO) COMPLEX, Proto-oncogene serine/threonine-protein kinase Pim-1, SULFATE ION | | Authors: | Maksimoska, J, Filippakopoulos, P, Knapp, S, Meggers, E. | | Deposit date: | 2008-01-09 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Similar biological activities of two isostructural ruthenium and osmium complexes.
Chemistry, 14, 2008
|
|
6EQ9
 
 | | Crystal structure of JNK3 in complex with AMP-PCP | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Macedo, J.T, Stehle, T, Blaum, B.S. | | Deposit date: | 2017-10-12 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Optimization of a Pyridinylimidazole Scaffold: Shifting the Selectivity from p38 alpha Mitogen-Activated Protein Kinase to c-Jun N-Terminal Kinase 3.
ACS Omega, 3, 2018
|
|
3GPB
 
 | |
3Q10
 
 | | Pantoate-beta-alanine ligase from Yersinia pestis | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Osipiuk, J, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Pantoate-beta-alanine ligase from Yersinia pestis.
To be Published
|
|
1JGT
 
 | | CRYSTAL STRUCTURE OF BETA-LACTAM SYNTHETASE | | Descriptor: | BETA-LACTAM SYNTHETASE, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, GLYCEROL, ... | | Authors: | Miller, M.T, Bachmann, B.O, Townsend, C.A, Rosenzweig, A.C. | | Deposit date: | 2001-06-26 | | Release date: | 2001-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of beta-lactam synthetase reveals how to synthesize antibiotics instead of asparagine.
Nat.Struct.Biol., 8, 2001
|
|
3W2W
 
 | | Crystal structure of the Cmr2dHD-Cmr3 subcomplex bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cmr subunit Cmr2, CRISPR system Cmr subunit Cmr3, ... | | Authors: | Numata, T, Osawa, T. | | Deposit date: | 2012-12-06 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Cmr2-Cmr3 Subcomplex in the CRISPR-Cas RNA Silencing Effector Complex.
J.Mol.Biol., 425, 2013
|
|
3KXH
 
 | | Crystal structure of Z. mays CK2 kinase alpha subunit in complex with the inhibitor (2-dymethylammino-4,5,6,7-tetrabromobenzoimidazol-1yl-acetic acid (K66) | | Descriptor: | Casein kinase II subunit alpha, DI(HYDROXYETHYL)ETHER, [4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzimidazol-1-yl]acetic acid | | Authors: | Papinutto, E, Franchin, C, Battistutta, R. | | Deposit date: | 2009-12-03 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | ATP site-directed inhibitors of protein kinase CK2: an update.
Curr Top Med Chem, 11, 2011
|
|
6ORU
 
 | |
3KXN
 
 | | Crystal structure of Z. mays CK2 kinase alpha subunit in complex with the inhibitor tetraiodobenzimidazole (K88) | | Descriptor: | 4,5,6,7-tetraiodo-1H-benzimidazole, Casein kinase II subunit alpha | | Authors: | Papinutto, E, Franchin, C, Battistutta, R. | | Deposit date: | 2009-12-03 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ATP site-directed inhibitors of protein kinase CK2: an update.
Curr Top Med Chem, 11, 2011
|
|
4XJ7
 
 | |
6O83
 
 | | S. pombe ubiquitin E1~ubiquitin-AMP tetrahedral intermediate mimic | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-{[(3-aminopropyl)sulfonyl]amino}-5'-deoxyadenosine, ... | | Authors: | Hann, Z.S, Lima, C.D. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.153 Å) | | Cite: | Structural basis for adenylation and thioester bond formation in the ubiquitin E1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3KXG
 
 | | Crystal structure of Z. mays CK2 kinase alpha subunit in complex with the inhibitor 3,4,5,6,7-pentabromo-1H-indazole (K64) | | Descriptor: | 3,4,5,6,7-pentabromo-1H-indazole, Casein kinase II subunit alpha | | Authors: | Papinutto, E, Franchin, C, Battistutta, R. | | Deposit date: | 2009-12-03 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | ATP site-directed inhibitors of protein kinase CK2: an update.
Curr Top Med Chem, 11, 2011
|
|
6P7P
 
 | | Structure of E. coli MS115-1 NucC, cAAA-bound form | | Descriptor: | CHLORIDE ION, Cyclic tri-AMP (5'-3' linked), E. coli MS115-1 NucC 2-241 | | Authors: | Ye, Q, Lau, R.K, Berg, K.R, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
4OKJ
 
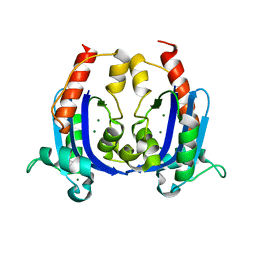 | | Crystal structure of RNase AS from M tuberculosis | | Descriptor: | 3'-5' exoribonuclease Rv2179c/MT2234.1, MAGNESIUM ION | | Authors: | Romano, M, van de Weerd, R, Brouwer, F.C.C, Roviello, G.N, Lacroix, R, Sparrius, M, van den Brink-van Stempvoort, G, Maaskant, J.J, van der Sar, A.M, Appelmelk, B.J, Geurtsen, J.J, Berisio, R. | | Deposit date: | 2014-01-22 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Function of RNase AS, a Polyadenylate-Specific Exoribonuclease Affecting Mycobacterial Virulence In Vivo
Structure, 22, 2014
|
|
4OKK
 
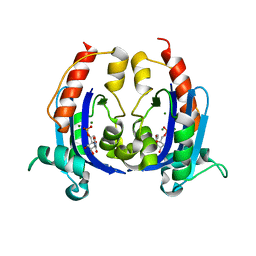 | | Crystal structure of RNase AS from M tuberculosis in complex with UMP | | Descriptor: | 3'-5' exoribonuclease Rv2179c/MT2234.1, MAGNESIUM ION, URIDINE-5'-MONOPHOSPHATE | | Authors: | Romano, M, van de Weerd, R, Brouwer, F.C.C, Roviello, G.N, Lacroix, R, Sparrius, M, van den Brink-van Stempvoort, G, Maaskant, J.J, van der Sar, A.M, Appelmelk, B.J, Geurtsen, J.J, Berisio, R. | | Deposit date: | 2014-01-22 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure and Function of RNase AS, a Polyadenylate-Specific Exoribonuclease Affecting Mycobacterial Virulence In Vivo
Structure, 22, 2014
|
|
7CVH
 
 | | Human Fructose-1,6-bisphosphatase 1 in complex with geranylgeranyl diphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Chen, Y, Zhang, J, Li, C, Cao, Y. | | Deposit date: | 2020-08-26 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The structural basis for GGPP activation on the enzymatic activities FBP1
To Be Published
|
|
