4AXG
 
 | | Structure of eIF4E-Cup complex | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, PROTEIN CUP | | Authors: | Kinkelin, K, Veith, K, Gruenwald, M, Bono, F. | | Deposit date: | 2012-06-12 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Minimal Eif4E-Cup Complex Revelas a General Mechanism of Eif4E Regulation in Translational Repression
RNA, 18, 2012
|
|
8VK3
 
 | |
6VI9
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - an alkylperoxo bound structure | | Descriptor: | (5R,6Z)-5-(hydroperoxy-kappaO)-5-(hydroxy-kappaO)-6-iminocyclohexa-1,3-diene-1-carboxylato(2-)iron, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VYR
 
 | | Escherichia coli transcription-translation complex A1 (TTC-A1) containing an 18 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
3ZNU
 
 | | Crystal structure of ClcF in crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION, ... | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
8W7M
 
 | | Yeast replisome in state V | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (71-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
2AEH
 
 | | Focal adhesion kinase 1 | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Ceccarelli, D.F, Song, H.K, Poy, F, Schaller, M.D, Eck, M.J. | | Deposit date: | 2005-07-22 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal Structure of the FERM Domain of Focal Adhesion Kinase
J.Biol.Chem., 281, 2006
|
|
2A1I
 
 | | Crystal Structure of the Central Domain of Human ERCC1 | | Descriptor: | DNA excision repair protein ERCC-1, MERCURY (II) ION | | Authors: | Tsodikov, O.V, Enzlin, J.H, Scharer, O.D, Ellenberger, T. | | Deposit date: | 2005-06-20 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and DNA binding functions of ERCC1, a subunit of the DNA structure-specific endonuclease XPF-ERCC1.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6W7W
 
 | | 30S-Inactive-low-Mg2+ Class B | | Descriptor: | 16S rRNA, 30S ribosomal protein S12, 30S ribosomal protein S15, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-19 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
6VYT
 
 | | Escherichia coli transcription-translation complex A2 (TTC-A2) containing a 15 nt long mRNA spacer, NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H, Su, M. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
2ADA
 
 | |
6VYY
 
 | | Escherichia coli transcription-translation complex C5 (TTC-C5) containing mRNA with a 21 nt long spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
2AJB
 
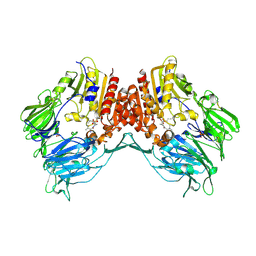 | | Porcine dipeptidyl peptidase IV (CD26) in complex with the tripeptide tert-butyl-Gly-L-Pro-L-Ile (tBu-GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-methyl-L-valyl-L-prolyl-L-isoleucine, ... | | Authors: | Engel, M, Hoffmann, T, Manhart, S, Heiser, U, Chambre, S, Huber, R, Demuth, H.U, Bode, W. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rigidity and flexibility of dipeptidyl peptidase IV: crystal structures of and docking experiments with DPIV.
J.Mol.Biol., 355, 2006
|
|
6VYW
 
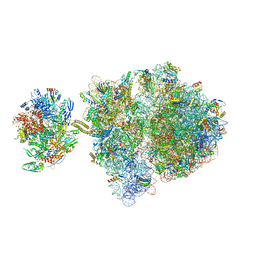 | | Escherichia coli transcription-translation complex C3 (TTC-C3) containing mRNA with a 27 nt long spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
8WPL
 
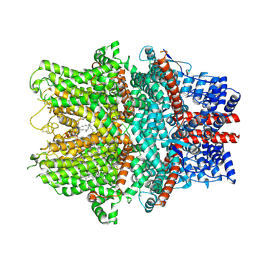 | | Cryo-EM structure of the human TRPC1/C4 heteromer | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Won, J, Jeong, H, Lee, H.H. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Cryo-EM structure of the heteromeric TRPC1/TRPC4 channel.
Nat.Struct.Mol.Biol., 2024
|
|
3ZLD
 
 | |
2C3L
 
 | | Identification of a buried pocket for potent and selective inhibition of Chk1: prediction and verification | | Descriptor: | 3-(1H-BENZIMIDAZOL-2-YL)-1H-INDAZOLE, SERINE/THREONINE-PROTEIN KINASE CHK1, SULFATE ION | | Authors: | Foloppe, N, Fisher, L.M, Francis, G, Howes, R, Kierstan, P, Potter, A. | | Deposit date: | 2005-10-10 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of a Buried Pocket for Potent and Selective Inhibition of Chk1: Prediction and Verification.
Bioorg.Med.Chem., 14, 2006
|
|
2V98
 
 | | Structure of the complex of TcAChE with 1-(2-nitrophenyl)-2,2,2- trifluoroethyl-arsenocholine after a 9 seconds annealing to room temperature, during the first 5 seconds of which laser irradiation at 266nm took place | | Descriptor: | 1-(2-nitrophenyl)-2,2,2-trifluoroethyl]-arsenocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Colletier, J.-P, Sanson, B, Royant, A, Specht, A, Nachon, F, Masson, P, Zaccai, G, Sussman, J.L, Goeldner, M, Silman, I, Bourgeois, D, Weik, M. | | Deposit date: | 2007-08-22 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Use of a 'Caged' Analog to Study Traffic of Choline within Acetylcholinesterase by Kinetic Crystallography
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4C90
 
 | | Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility | | Descriptor: | ALPHA-GLUCURONIDASE GH115, SODIUM ION | | Authors: | Rogowski, A, Basle, A, Farinas, C.S, Solovyova, A, Mortimer, J.C, Dupree, P, Gilbert, H.J, Bolam, D.N. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Evidence that Gh115 Alpha-Glucuronidase Activity is Dependent on Conformational Flexibility
J.Biol.Chem., 289, 2014
|
|
2C6L
 
 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | 4-({5-[(4-AMINOCYCLOHEXYL)AMINO][1,2,4]TRIAZOLO[1,5-A]PYRIMIDIN-7-YL}AMINO)BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-10 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Triazolo[1,5-A]Pyrimidines as Novel Cdk2 Inhibitors: Protein Structure-Guided Design and Sar.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8J07
 
 | |
8WPM
 
 | | Cryo-EM structure of the human TRPC1/C4 heteromer in complex with Pico145 | | Descriptor: | 7-[(4-chlorophenyl)methyl]-3-methyl-1-(3-oxidanylpropyl)-8-[3-(trifluoromethyloxy)phenoxy]purine-2,6-dione, CALCIUM ION, Short transient receptor potential channel 1, ... | | Authors: | Won, J, Jeong, H, Lee, H.H. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Cryo-EM structure of the heteromeric TRPC1/TRPC4 channel.
Nat.Struct.Mol.Biol., 2024
|
|
8WPN
 
 | | Cryo-EM structure of the human TRPC4 in lipid nanodiscs | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Won, J, Jeong, H, Lee, H.H. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Cryo-EM structure of the heteromeric TRPC1/TRPC4 channel.
Nat.Struct.Mol.Biol., 2024
|
|
8J7R
 
 | | Cryo-EM structure of the J-K-St region of EMCV IRES in complex with eIF4G-HEAT1 and eIF4A (J-K-St/eIF4G focused) | | Descriptor: | Eukaryotic translation initiation factor 4 gamma 1, IRES RNA (J-K-St), MAGNESIUM ION | | Authors: | Suzuki, H, Fujiyoshi, Y, Imai, S, Shimada, I. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Dynamically regulated two-site interaction of viral RNA to capture host translation initiation factor.
Nat Commun, 14, 2023
|
|
2V9V
 
 | | Crystal Structure of Moorella thermoacetica SelB(377-511) | | Descriptor: | CHLORIDE ION, SELENOCYSTEINE-SPECIFIC ELONGATION FACTOR, SODIUM ION | | Authors: | Ganichkin, O, Wahl, M.C. | | Deposit date: | 2007-08-27 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Conformational Switches in Winged-Helix Domains 1 and 2 of Bacterial Translation Elongation Factor Selb.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
