5LP1
 
 | |
7BOI
 
 | | Bacterial 30S ribosomal subunit assembly complex state F (multibody refinement for body domain of 30S ribosome) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7BOF
 
 | | Bacterial 30S ribosomal subunit assembly complex state I (body domain) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
8TSM
 
 | |
5J5K
 
 | | CRYSTAL STRUCTURE OF AFMP4P IN COMPLEX WITH PALMITIC ACID | | Descriptor: | PALMITIC ACID, Uncharacterized protein, alpha-D-mannopyranose | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2016-04-03 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Novel Class Of Virulence Factors In Penicillium Marneffei And Aspergillus Fumigatus Enhances Intracellular Survival In Monocytes By Arachidonic Acid Binding
To Be Published
|
|
5XRL
 
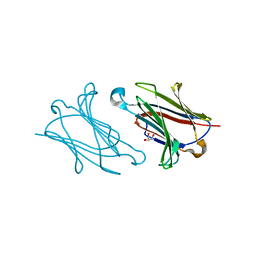 | |
4ASC
 
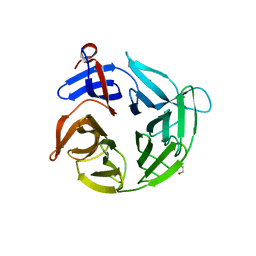 | | Crystal structure of the Kelch domain of human KBTBD5 | | Descriptor: | 1,2-ETHANEDIOL, KELCH REPEAT AND BTB DOMAIN-CONTAINING PROTEIN 5 | | Authors: | Canning, P, Ayinampudi, V, Krojer, T, Strain-Damerell, C, Raynor, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
6GUR
 
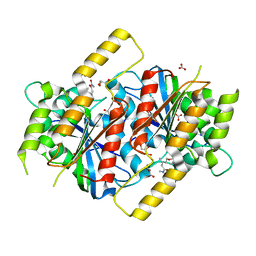 | | Siderophore hydrolase EstB from Aspergillus fumigatus in complex with TAFC | | Descriptor: | (~{Z})-5-[(1~{S},2~{S})-2-acetamido-1-oxidanyl-5-[oxidanyl(propanoyl)amino]pentoxy]-~{N},3-dimethyl-~{N}-oxidanyl-pent-2-enamide, CARBONATE ION, FE (III) ION, ... | | Authors: | Ecker, F, Haas, H, Groll, M, Huber, E.M. | | Deposit date: | 2018-06-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iron Scavenging in Aspergillus Species: Structural and Biochemical Insights into Fungal Siderophore Esterases.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5NGY
 
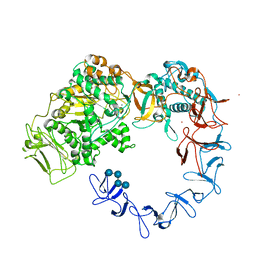 | | Crystal structure of Leuconostoc citreum NRRL B-1299 dextransucrase DSR-M | | Descriptor: | CALCIUM ION, DSR-M glucansucrase inactive mutant E715Q, PRASEODYMIUM ION, ... | | Authors: | Claverie, M, Cioci, G, Remaud-simeon, M, Moulis, C, Tranier, S, Vuillemin, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Investigations on the Determinants Responsible for Low Molar Mass Dextran Formation by DSR-M Dextransucrase
Acs Catalysis, 2017
|
|
5K6E
 
 | |
6H9B
 
 | | 1,1-Diheterocyclic Ethylenes Derived from Quinaldine and Carbazole as New Tubulin Polymerization Inhibitors: Synthesis, Metabolism, and Biological Evaluation | | Descriptor: | 9-methyl-3-[1-(2-methylquinolin-4-yl)ethenyl]carbazole, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Varela, P.F, Gigant, B. | | Deposit date: | 2018-08-03 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 1,1-Diheterocyclic Ethylenes Derived from Quinaldine and Carbazole as New Tubulin-Polymerization Inhibitors: Synthesis, Metabolism, and Biological Evaluation.
J. Med. Chem., 62, 2019
|
|
5WR0
 
 | | Huisgen cycloaddition for PPARg-LBD labeling by soaking method | | Descriptor: | (E)-N-[(3E)-2-oxo-16-(8-{6-[(trifluoroacetyl)amino]hexanoyl}-8,9-dihydro-1H-dibenzo[b,f][1,2,3]triazolo[4,5-d]azocin-1-yl)hexadec-3-en-1-ylidene]glycine, Peroxisome proliferator-activated receptor gamma | | Authors: | Kojima, H, Itoh, T, Yamamoto, K. | | Deposit date: | 2016-11-29 | | Release date: | 2017-11-22 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | On-site reaction for PPAR gamma modification using a specific bifunctional ligand
Bioorg. Med. Chem., 25, 2017
|
|
6OCJ
 
 | | Crystal Structure of Equine Serum Albumin in Complex with Suprofen | | Descriptor: | (2S)-2-[4-(thiophene-2-carbonyl)phenyl]propanoic acid, ACETATE ION, MALONATE ION, ... | | Authors: | Sekula, B, Zielinski, K, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-03-24 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural investigations of stereoselective profen binding by equine and leporine serum albumins.
Chirality, 32, 2020
|
|
3I4X
 
 | | Crystal structure of the dimethylallyl tryptophan synthase FgaPT2 from Aspergillus fumigatus in complex with Trp and DMSPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, GLYCEROL, TRYPTOPHAN, ... | | Authors: | Schall, C, Zocher, G, Stehle, T. | | Deposit date: | 2009-07-03 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6GL6
 
 | | apo [FeFe]-hydrogenase HydA1 from Chlamydomonas reinhardtii, variant C377H | | Descriptor: | CHLORIDE ION, Fe-hydrogenase, IRON/SULFUR CLUSTER, ... | | Authors: | Kertess, L, Happe, T, Hofmann, E. | | Deposit date: | 2018-05-23 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | His-Ligation to the [4Fe-4S] Subcluster Tunes the Catalytic Bias of [FeFe] Hydrogenase.
J.Am.Chem.Soc., 141, 2019
|
|
6OCI
 
 | | Crystal Structure of Equine Serum Albumin in Complex with Ibuprofen | | Descriptor: | FORMIC ACID, IBUPROFEN, SUCCINIC ACID, ... | | Authors: | Sekula, B, Zielinski, K, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-03-24 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural investigations of stereoselective profen binding by equine and leporine serum albumins.
Chirality, 32, 2020
|
|
4ZCG
 
 | | Crystal Structure of human GGT1 in complex with Glutamate (with all atoms of glutamate) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Terzyan, S, Hanigan, M. | | Deposit date: | 2015-04-15 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Human gamma-Glutamyl Transpeptidase 1: STRUCTURES OF THE FREE ENZYME, INHIBITOR-BOUND TETRAHEDRAL TRANSITION STATES, AND GLUTAMATE-BOUND ENZYME REVEAL NOVEL MOVEMENT WITHIN THE ACTIVE SITE DURING CATALYSIS.
J.Biol.Chem., 290, 2015
|
|
2PHH
 
 | |
4GDE
 
 | |
6H4D
 
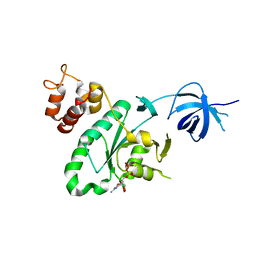 | | Crystal structure of RsgA from Pseudomonas aeruginosa | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Small ribosomal subunit biogenesis GTPase RsgA, ZINC ION | | Authors: | Rocchio, S, Santorelli, D, Travaglini-Allocatelli, C, Federici, L, Di Matteo, A. | | Deposit date: | 2018-07-20 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional investigation of the Small Ribosomal Subunit Biogenesis GTPase A (RsgA) from Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
7Y4K
 
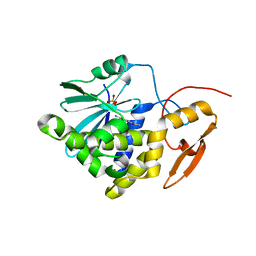 | | Crystal structure of Ricin A chain bound with N2-(2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)glycyl-L-phenylalanyl-N6-((benzyloxy)carbonyl)-L-ornitine | | Descriptor: | (2S)-2-[[(2S)-2-[2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]ethanoylamino]-3-phenyl-propanoyl]amino]-5-(phenylmethoxycarbonylamino)pentanoic acid, Ricin A chain, SULFATE ION | | Authors: | Katakura, S, Goto, M, Ohba, T, Kawata, R, Nagatsu, K, Higashi, S, Matsumoto, K, Kurisu, K, Ohtsuka, K, Saito, R. | | Deposit date: | 2022-06-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pterin-based small molecule inhibitor capable of binding to the secondary pocket in the active site of ricin-toxin A chain.
Plos One, 17, 2022
|
|
7Y4M
 
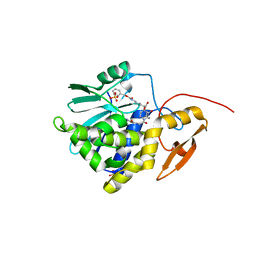 | | Crystal structure of Ricin A chain bound with N2-(2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)glycyl-L-phenylalanyl-N6-((benzyloxy)carbonyl)-L-lysine | | Descriptor: | (2S)-2-[[(2S)-2-[2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]ethanoylamino]-3-phenyl-propanoyl]amino]-6-(phenylmethoxycarbonylamino)hexanoic acid, Ricin A chain, SULFATE ION | | Authors: | Katakura, S, Goto, M, Ohba, T, Kawata, R, Nagatsu, K, Higashi, S, Matsumoto, K, Kurisu, K, Ohtsuka, K, Saito, R. | | Deposit date: | 2022-06-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Pterin-based small molecule inhibitor capable of binding to the secondary pocket in the active site of ricin-toxin A chain.
Plos One, 17, 2022
|
|
3UP3
 
 | | Nuclear receptor DAF-12 from hookworm Ancylostoma ceylanicum in complex with (25S)-cholestenoic acid | | Descriptor: | (8alpha,10alpha,25S)-3-hydroxycholesta-3,5-dien-26-oic acid, 1,2-ETHANEDIOL, Nuclear receptor coactivator 2, ... | | Authors: | Zhi, X, Zhou, X.E, Melcher, K, Motola, D.L, Gelmedin, V, Hawdon, J, Kliewer, S.A, Mangelsdorf, D.J, Xu, H.E. | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Conservation of Ligand Binding Reveals a Bile Acid-like Signaling Pathway in Nematodes.
J.Biol.Chem., 287, 2012
|
|
3KHQ
 
 | | Crystal structure of murine Ig-beta (CD79b) in the monomeric form | | Descriptor: | B-cell antigen receptor complex-associated protein beta chain, CITRATE ANION, GLUTATHIONE, ... | | Authors: | Radaev, S, Sun, P.D. | | Deposit date: | 2009-10-30 | | Release date: | 2010-08-25 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Studies of Igalphabeta and Its Assembly with the B Cell Antigen Receptor.
Structure, 18, 2010
|
|
6IK6
 
 | | Crystal structure of Tomato beta-galactosidase (TBG) 4 with beta-1,4-galactobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Matsuyama, K, Nakae, S, Igarashi, K, Tada, T, Ishimaru, M. | | Deposit date: | 2018-10-15 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.791 Å) | | Cite: | Substrate-recognition mechanism of tomato beta-galactosidase 4 using X-ray crystallography and docking simulation.
Planta, 252, 2020
|
|
