5P4V
 
 | |
5P6T
 
 | |
5P59
 
 | |
5P77
 
 | |
1HTG
 
 | |
1WMZ
 
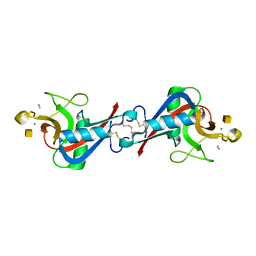 | | Crystal Structure of C-type Lectin CEL-I complexed with N-acetyl-D-galactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Sugawara, H, Kusunoki, M, Kurisu, G, Fujimoto, T, Aoyagi, H, Hatakeyama, T. | | Deposit date: | 2004-07-22 | | Release date: | 2004-09-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characteristic Recognition of N-Acetylgalactosamine by an Invertebrate C-type Lectin, CEL-I, Revealed by X-ray Crystallographic Analysis
J.Biol.Chem., 279, 2004
|
|
5P5P
 
 | |
5P7I
 
 | |
1HJ8
 
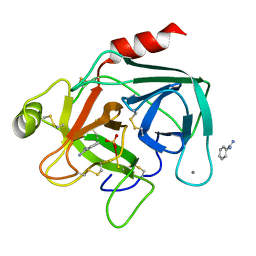 | | 1.00 AA Trypsin from Atlantic Salmon | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Leiros, H.-K.S, Mcsweeney, S.M, Smalas, A.O. | | Deposit date: | 2001-01-09 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic Resolution Structure of Trypsin Provide Insight Into Structural Radiation Damage
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3EZR
 
 | | CDK-2 with indazole inhibitor 17 bound at its active site | | Descriptor: | 3-methoxy-4-{3-[4-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2 | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-10-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5P65
 
 | |
5P7X
 
 | |
2AL7
 
 | | Structure Of Human ADP-Ribosylation Factor-Like 10C | | Descriptor: | ADP-ribosylation factor-like 10C, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Ismail, S, Dimov, S, Atanassova, A, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-04 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | GTP-like conformation of GDP-bound ARL10C GTPase
To be Published
|
|
5P6J
 
 | |
1HVO
 
 | |
5P8C
 
 | |
5P8P
 
 | |
5P6X
 
 | |
1HPV
 
 | |
1HI2
 
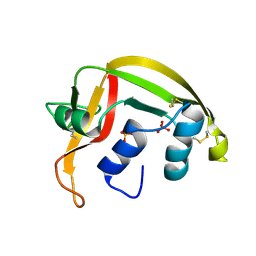 | | Eosinophil-derived Neurotoxin (EDN) - Sulphate Complex | | Descriptor: | EOSINOPHIL-DERIVED NEUROTOXIN, SULFATE ION | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
2AOG
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a substrate analog P2-NC | | Descriptor: | ACETIC ACID, GLYCEROL, HIV-1 PROTEASE (RETROPEPSIN), ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
5P7B
 
 | |
2AOZ
 
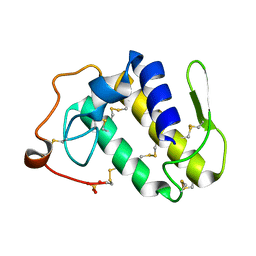 | | Crystal structure of the myotoxin-II from Atropoides nummifer venom | | Descriptor: | Phospholipase A2 homolog, SULFATE ION | | Authors: | Melo, C.C, Murakami, M.T, Angulo, Y, Lomonte, B, Arni, R.K. | | Deposit date: | 2005-08-15 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of myotoxin II, a catalytically inactive Lys49 phospholipase A2 homologue from Atropoides nummifer venom.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
5P7P
 
 | |
1HI5
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
