5IFN
 
 | |
5IFP
 
 | | STRUCTURE OF BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rico-Diaz, A, Ramirez-Escudero, M, Vizoso Vazquez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2016-02-26 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural features of Aspergillus niger beta-galactosidase define its activity against glycoside linkages.
FEBS J., 284, 2017
|
|
5IFT
 
 | | STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 3-b-Galactopyranosyl glucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Rico-Diaz, A, Ramirez-Escudero, M, Vizoso Vazquez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2016-02-26 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural features of Aspergillus niger beta-galactosidase define its activity against glycoside linkages.
FEBS J., 284, 2017
|
|
8IJG
 
 | | Crystal structure of alcohol dehydrogenase M5 from Burkholderia gladioli with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative short-chain dehydrogenases/reductase family protein | | Authors: | Han, X, Mei, Z.L, Liu, W.D, Sun, Z.T, Ma, J.A. | | Deposit date: | 2023-02-27 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of alcohol dehydrogenase from Burkholderia gladioli with NADP
To Be Published
|
|
5IG1
 
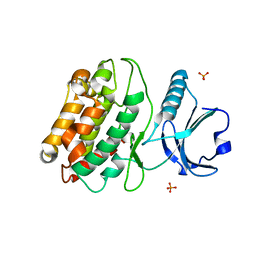 | | Crystal structure of S. rosetta CaMKII kinase domain | | Descriptor: | CAMK/CAMK2 protein kinase, PHOSPHATE ION | | Authors: | Bhattacharyya, M, Gee, C.L, Barros, T, Kuriyan, J. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
8FOV
 
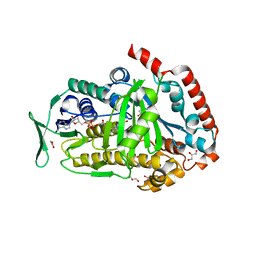 | | AbeH (Tryptophan-5-halogenase) bound to FAD and Cl | | Descriptor: | ACETATE ION, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ashaduzzaman, M, Bellizzi, J.J. | | Deposit date: | 2023-01-03 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystallographic and thermodynamic evidence of negative cooperativity of flavin and tryptophan binding in the flavin-dependent halogenases AbeH and BorH.
Biorxiv, 2023
|
|
8ILU
 
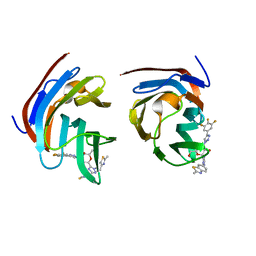 | | Crystal structure of mouse Galectin-3 in complex with small molecule inhibitor | | Descriptor: | (2R,3R,4R,5R,6S)-2-(hydroxymethyl)-6-[2-(2-methyl-1,3-benzothiazol-6-yl)-1,2,4-triazol-3-yl]-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3, SODIUM ION, ... | | Authors: | Kumar, A, Jinal, S, Raman, S, Ghosh, K. | | Deposit date: | 2023-03-04 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of Benzothiazole Derived Monosaccharides as Potent, Selective, and Orally Bioavailable Inhibitors of Human and Mouse Galectin-3
To Be Published
|
|
8ID1
 
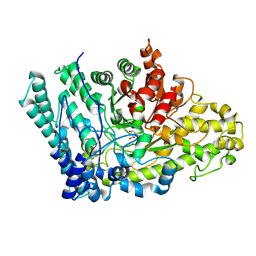 | |
5IH3
 
 | | Crystal structure of mouse CARM1 in complex with SAH at 1.8 Angstroms resolution | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with SAH at 1.8 Angstroms resolution
To Be Published
|
|
8FZA
 
 | | Class I type III preQ1 riboswitch from E. coli | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, MANGANESE (II) ION, PreQ1 Riboswitch (30-MER) | | Authors: | Wedekind, J.E, Schroeder, G.M, Jenkins, J.L. | | Deposit date: | 2023-01-27 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function analysis of a type III preQ 1 -I riboswitch from Escherichia coli reveals direct metabolite sensing by the Shine-Dalgarno sequence.
J.Biol.Chem., 299, 2023
|
|
8FII
 
 | | Wild type APOBEC3A in complex with TT(FdZ)-hairpin inhibitor (crystal form 1) | | Descriptor: | DNA (5'-D(P*GP*CP*GP*CP*TP*TP*(UFP)P*GP*CP*GP*C)-3'), DNA dC->dU-editing enzyme APOBEC-3A, PHOSPHATE ION, ... | | Authors: | Harjes, S, Jameson, G.B, Harjes, E, Filichev, V.V, Kurup, H.M. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | DNA-based inhibitors restrict mutagenic activity of APOBEC3 in cells
To Be Published
|
|
8FIJ
 
 | | Wild type APOBEC3A in complex with TT(FdZ)-hairpin inhibitor (crystal form 2) | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*TP*TP*(UFP)P*GP*CP*GP*CP*T)-3'), DNA dC->dU-editing enzyme APOBEC-3A, PHOSPHATE ION, ... | | Authors: | Harjes, S, Jameson, G.B, Harjes, E, Filichev, V.V, Kurup, H.M. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | DNA-based inhibitors restrict mutagenic activity of APOBEC3 in cells
To Be Published
|
|
5IG4
 
 | | Crystal structure of N. vectensis CaMKII-A hub | | Descriptor: | GLYCEROL, Predicted protein | | Authors: | Bhattacharyya, M, Pappireddi, N, Gee, C.L, Barros, T, Kuriyan, J. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
8FIM
 
 | | Structure of APOBEC3A (E72A inactive mutant) in complex with TTC-hairpin DNA substrate | | Descriptor: | CHLORIDE ION, DNA (5'-D(*TP*GP*CP*GP*CP*TP*TP*CP*GP*CP*GP*CP*T)-3'), DNA dC->dU-editing enzyme APOBEC-3A, ... | | Authors: | Harjes, S, Jameson, G.B, Harjes, E, Filichev, V.V, Kurup, H.M. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | DNA-based inhibitors restrict mutagenic activity of APOBEC3 in cells
To Be Published
|
|
5IGQ
 
 | |
8IN0
 
 | |
8FEW
 
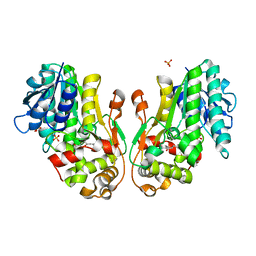 | | Flavanone 4-Reductase from Sorghum bicolor-naringenin complex | | Descriptor: | 3-deoxyanthocyanidin synthase, NARINGENIN, SULFATE ION | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
5IK5
 
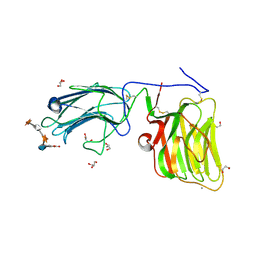 | | Laminin A2LG45 C-form, G6/7 bound. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-hydroxy-4-methyl-2H-chromen-2-one, CALCIUM ION, ... | | Authors: | Briggs, D.C, Hohenester, E, Campbell, K.P. | | Deposit date: | 2016-03-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural basis of laminin binding to the LARGE glycans on dystroglycan.
Nat.Chem.Biol., 12, 2016
|
|
5IKD
 
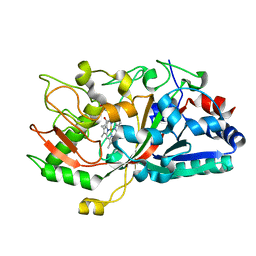 | |
5IIN
 
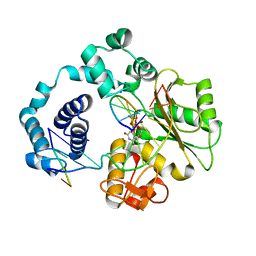 | | Crystal structure of the pre-catalytic ternary extension complex of DNA polymerase lambda with an 8-oxo-dG:dC base-pair | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*(8OG)P*TP*AP*CP*TP*G)-3'), ... | | Authors: | Burak, M.J, Guja, K.E, Garcia-Diaz, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A fidelity mechanism in DNA polymerase lambda promotes error-free bypass of 8-oxo-dG.
Embo J., 35, 2016
|
|
5IKW
 
 | | Crystal Structure of BMP-2-inducible kinase in complex with an Indazole inhibitor | | Descriptor: | BMP-2-inducible protein kinase, N-(6-{3-[(cyclopropylsulfonyl)amino]phenyl}-1H-indazol-3-yl)cyclopropanecarboxamide | | Authors: | Counago, R.M, Sorrell, F.J, Krojer, T, Savitsky, P, Elkins, J.M, Axtman, A, Drewry, D, Wells, C, Zhang, C, Zuercher, W, Willson, T.M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Arruda, P, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-04 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal Structure of BMP-2-inducible kinase in complex with a 3-acylaminoindazole inhibitor GSK3236425A
To Be Published
|
|
5IL1
 
 | | Crystal structure of SAM-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
8INL
 
 | | LSD1 in complex with S2172 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-phenylmethoxy-phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | LSD1 in complex with S2172
To Be Published
|
|
8IN7
 
 | | Crystal structure of UGT74AN3-UDP | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Feng, L, Wei, H. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of UGT74AN3-UDP
To Be Published
|
|
5ILI
 
 | | Tobacco 5-epi-aristolochene synthase with CAPSO buffer molecule and Mg2+ ions | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 5-epi-aristolochene synthase, ... | | Authors: | Koo, H.J, Louie, G.V, Xu, Y, Bowman, M, Noel, J.P. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small-molecule buffer components can directly affect terpene-synthase activity by interacting with the substrate-binding site of the enzyme
To Be Published
|
|
