5T56
 
 | |
6TYW
 
 | |
6KJN
 
 | | The microtubule-binding domains of yeast cytoplasmic dynein in the high affinity state | | Descriptor: | Dynein heavy chain, cytoplasmic | | Authors: | Nishida, N, Komori, Y, Takarada, O, Watanabe, A, Tamura, S, Kubo, S, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
5WPY
 
 | |
6TYZ
 
 | |
5WUZ
 
 | | Morintides mO1 | | Descriptor: | Morintide mO1 | | Authors: | Xiao, T, Tam, J.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Morintides: 8C-Hevein-like peptides without a protein cargo from the drumstick tree Moringa oleifera
To Be Published
|
|
1IXT
 
 | | Structure of a Novel P-Superfamily Spasmodic Conotoxin Reveals an Inhibitory Cystine Knot Motif | | Descriptor: | spasmodic protein tx9a-like protein | | Authors: | Miles, L.A, Dy, C.Y, Nielsen, J, Barnham, K.J, Hinds, M.G, Olivera, B.M, Bulaj, G, Norton, R.S. | | Deposit date: | 2002-07-04 | | Release date: | 2003-01-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of a Novel P-Superfamily Spasmodic Conotoxin Reveals an Inhibitory Cystine Knot Motif
J.Biol.Chem., 277, 2002
|
|
6K84
 
 | | Structure of anti-prion RNA aptamer | | Descriptor: | RNA (25-MER) | | Authors: | Mashima, T, Lee, J.H, Hayashi, T, Nagata, T, Kinoshita, M, Katahira, M. | | Deposit date: | 2019-06-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Development and structural determination of an anti-PrPCaptamer that blocks pathological conformational conversion of prion protein.
Sci Rep, 10, 2020
|
|
1IV0
 
 | |
6TYV
 
 | |
1IEN
 
 | | SOLUTION STRUCTURE OF TIA | | Descriptor: | PROTEIN TIA | | Authors: | Sharpe, I.A, Gehrmann, J, Loughnan, M.L, Thomas, L, Adams, D.A, Atkins, A, Palant, E, Craik, D.J, Adams, D.J, Alewood, P.F, Lewis, R.J. | | Deposit date: | 2001-04-10 | | Release date: | 2002-04-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Two new classes of conopeptides inhibit the alpha1-adrenoceptor and noradrenaline transporter.
Nat.Neurosci., 4, 2001
|
|
1IFW
 
 | | SOLUTION STRUCTURE OF C-TERMINAL DOMAIN OF POLY(A) BINDING PROTEIN FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | POLYADENYLATE-BINDING PROTEIN, CYTOPLASMIC AND NUCLEAR | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Sprules, T, Ekiel, I, Gehring, K. | | Deposit date: | 2001-04-13 | | Release date: | 2002-07-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the orphan PABC domain from Saccharomyces cerevisiae poly(A)-binding protein.
J.Biol.Chem., 277, 2002
|
|
1IGV
 
 | | BOVINE CALBINDIN D9K BINDING MN2+ | | Descriptor: | MANGANESE (II) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN, INTESTINAL | | Authors: | Andersson, E.M. | | Deposit date: | 2001-04-18 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the negative allostery between Ca(2+)- and Mg(2+)-binding in the intracellular Ca(2+)-receptor calbindin D9k.
Protein Sci., 6, 1997
|
|
6TYX
 
 | |
1IU2
 
 | | The first PDZ domain of PSD-95 | | Descriptor: | PSD-95 | | Authors: | Long, J.-F, Tochio, H, Wang, P, Sala, C, Niethammer, M, Sheng, M, Zhang, M. | | Deposit date: | 2002-02-19 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Supramodular structure and synergistic target binding of the N-terminal tandem PDZ domains of PSD-95
J.MOL.BIOL., 327, 2003
|
|
1IG5
 
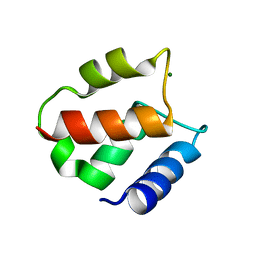 | | BOVINE CALBINDIN D9K BINDING MG2+ | | Descriptor: | MAGNESIUM ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN, INTESTINAL | | Authors: | Andersson, E.M, Svensson, L.A. | | Deposit date: | 2001-04-17 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the negative allostery between Ca(2+)- and Mg(2+)-binding in the intracellular Ca(2+)-receptor calbindin D9k.
Protein Sci., 6, 1997
|
|
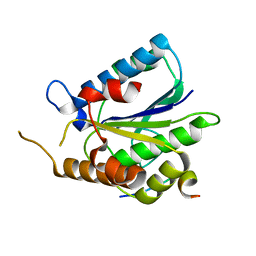
6TYU
 
 | |
1BDW
 
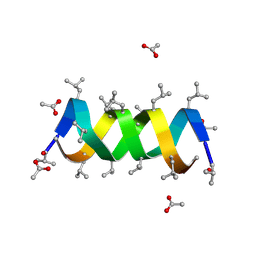 | |
2RPN
 
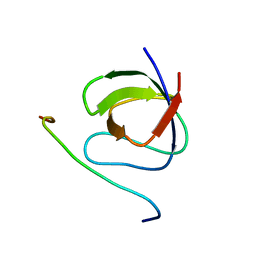 | | A crucial role for high intrinsic specificity in the function of yeast SH3 domains | | Descriptor: | Actin-binding protein, Actin-regulating kinase 1 | | Authors: | Stollar, E.J, Garcia, B, Chong, A, Forman-Kay, J, Davidson, A. | | Deposit date: | 2008-06-12 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural, functional, and bioinformatic studies demonstrate the crucial role of an extended peptide binding site for the SH3 domain of yeast Abp1p
J.Biol.Chem., 284, 2009
|
|
1J3C
 
 | |
1J3X
 
 | |
6TYT
 
 | |
6TVM
 
 | | LEDGF/p75 dimer (residues 345-467) | | Descriptor: | PC4 and SFRS1-interacting protein | | Authors: | Lux, V, Veverka, V. | | Deposit date: | 2020-01-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Molecular Mechanism of LEDGF/p75 Dimerization.
Structure, 28, 2020
|
|
6KXZ
 
 | |
1J0Q
 
 | | Solution Structure of Oxidized Bovine Microsomal Cytochrome b5 mutant V61H | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Wu, H, Huang, Z, Cao, C, Zhang, Q, Wang, Y.-H, Ma, J.-B, Xue, L.-L. | | Deposit date: | 2002-11-20 | | Release date: | 2003-08-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the oxidized bovine microsomal cytochrome b5 mutant V61H
Biochem.Biophys.Res.Commun., 307, 2003
|
|
