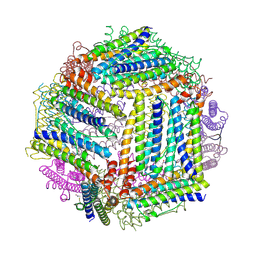
4ISP
 
 | |
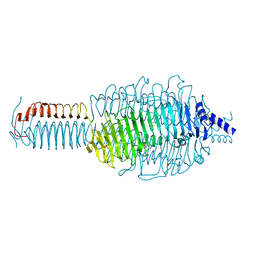
3GQ7
 
 | |
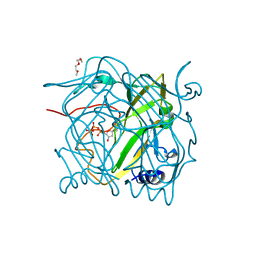
2QXX
 
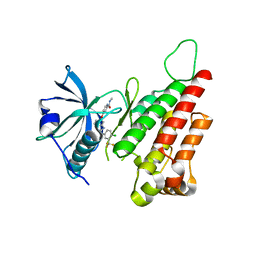
 | | Bifunctional dCTP deaminase: dUTPase from Mycobacterium tuberculosis in complex with dTTP | | Descriptor: | Deoxycytidine triphosphate deaminase, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Christophersen, S, Harris, P, Willemoes, M. | | Deposit date: | 2007-08-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of dTTP inhibition of the bifunctional dCTP deaminase:dUTPase encoded by Mycobacterium tuberculosis.
J.Mol.Biol., 376, 2008
|
|
2QU5
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a benzimidazole inhibitor | | Descriptor: | 4-[[2-[[4-chloro-3-(trifluoromethyl)phenyl]amino]-3H-benzimidazol-5-yl]oxy]-N-methyl-pyridine-2-carboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Kim, J.L, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2007-08-03 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Design, Synthesis, and Evaluation of Orally Active Benzimidazoles and Benzoxazoles as Vascular Endothelial Growth Factor-2 Receptor Tyrosine Kinase Inhibitors.
J.Med.Chem., 50, 2007
|
|
3QIL
 
 | |
2R0W
 
 | | PFA2 FAB complexed with Abeta1-8 | | Descriptor: | Amyloid beta peptide fragment, IgG2a Fab fragment heavy chain, Fd portion, ... | | Authors: | Gardberg, A.S, Dealwis, C. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-16 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Molecular basis for passive immunotherapy of Alzheimer's disease
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3HBL
 
 | | Crystal Structure of S. aureus Pyruvate Carboxylase T908A Mutant | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Tong, L, Yu, L.P.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A Symmetrical Tetramer for S. aureus Pyruvate Carboxylase in Complex with Coenzyme A.
Structure, 17, 2009
|
|
3QLZ
 
 | | Candida glabrata dihydrofolate reductase complexed with NADPH and 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-propylpyrimidine-2,4-diamine (UCP130B) | | Descriptor: | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-propylpyrimidine-2,4-diamine, MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Paulsen, J.L, Bendel, S.D, Anderson, A.C. | | Deposit date: | 2011-02-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structures of Candida albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical for Ligand Potency and Selectivity.
Chem.Biol.Drug Des., 78, 2011
|
|
4IZ8
 
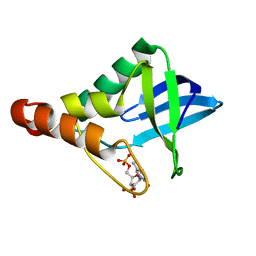 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS H8E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Bell-Upp, P.C, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-01-29 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS H8E at cryogenic temperature
To be Published
|
|
3QDE
 
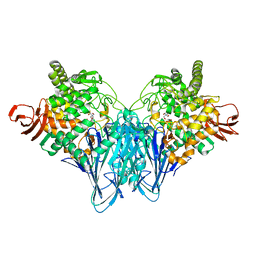 | | The structure of Cellobiose phosphorylase from Clostridium thermocellum in complex with phosphate | | Descriptor: | Cellobiose phosphorylase, PHOSPHATE ION, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM | | Authors: | Bianchetti, C.M, Elsen, N.L, Horn, M.K, Fox, B.G, Phillips Jr, G.N. | | Deposit date: | 2011-01-18 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of cellobiose phosphorylase from Clostridium thermocellum in complex with phosphate.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2R46
 
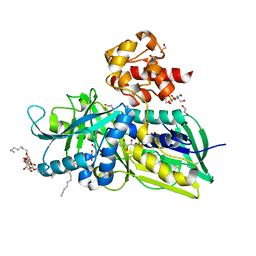 | | Crystal structure of Escherichia coli Glycerol-3-phosphate Dehydrogenase in complex with 2-phosphopyruvic acid. | | Descriptor: | 1,2-ETHANEDIOL, Aerobic glycerol-3-phosphate dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yeh, J.I, Du, S, Chinte, U. | | Deposit date: | 2007-08-30 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential
monotopic membrane enzyme involved in respiration and metabolism
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4J1M
 
 | |
2QEL
 
 | |
2R0J
 
 | | Crystal structure of the putative ubiquitin conjugating enzyme, PFE1350c, from Plasmodium falciparum | | Descriptor: | Ubiquitin carrier protein | | Authors: | Wernimont, A.K, Lew, J, Lin, Y.H, Hassanali, A, Kozieradzki, I, Zhao, Y, Schapira, M, Bochkarev, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-20 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the putative ubiquitin conjugating enzyme, PFE1350c, from Plasmodium falciparum.
To be Published
|
|
4ITL
 
 | | Crystal structure of LpxK from Aquifex aeolicus in complex with AMP-PCP at 2.1 angstrom resolution | | Descriptor: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Tetraacyldisaccharide 4'-kinase | | Authors: | Emptage, R.P, Pemble IV, C.W, York, J.D, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Mechanistic Characterization of the Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK Involved in Lipid A Biosynthesis.
Biochemistry, 52, 2013
|
|
3QR0
 
 | | Crystal Structure of S. officinalis PLC21 | | Descriptor: | CALCIUM ION, GLYCEROL, phospholipase C-beta (PLC-beta) | | Authors: | Lyon, A.M, Northup, J.K, Tesmer, J.J.G. | | Deposit date: | 2011-02-16 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An autoinhibitory helix in the C-terminal region of phospholipase C-beta mediates Galphaq activation.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3GOR
 
 | | Crystal structure of putative metal-dependent hydrolase APC36150 | | Descriptor: | NICKEL (II) ION, Putative metal-dependent hydrolase | | Authors: | Cooper, D.R, Grelewska, K, Derewenda, Z.S, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2009-03-19 | | Release date: | 2009-05-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.511 Å) | | Cite: | The structure of DinB from Geobacillus stearothermophilus: a representative of a unique four-helix-bundle superfamily.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3QIA
 
 | | Crystal structure of P-loop G237A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-26 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
2QO7
 
 | | Human EphA3 kinase and juxtamembrane region, dephosphorylated, AMP-PNP bound | | Descriptor: | BETA-MERCAPTOETHANOL, Ephrin receptor, GLYCEROL, ... | | Authors: | Davis, T, Walker, J.R, Newman, E.M, Mackenzie, F, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Autoregulation by the Juxtamembrane Region of the Human Ephrin Receptor Tyrosine Kinase A3 (EphA3).
Structure, 16, 2008
|
|
3QJ8
 
 | |
2QOI
 
 | | Human EphA3 kinase and juxtamembrane region, Y596F:Y602F double mutant | | Descriptor: | BETA-MERCAPTOETHANOL, Ephrin receptor | | Authors: | Davis, T, Walker, J.R, Newman, E.M, Mackenzie, F, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Autoregulation by the Juxtamembrane Region of the Human Ephrin Receptor Tyrosine Kinase A3 (EphA3).
Structure, 16, 2008
|
|
3GSO
 
 | | Crystal structure of the binary complex between HLA-A2 and HCMV NLV peptide | | Descriptor: | Beta-2-microglobulin, HCMV pp65 fragment 495-503 (NLVPMVATV), HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Saulquin, X, Reiser, J.-B, Debeaupuis, E, Echasserieau, K, Kissenpfennig, A, Legoux, F, Chouquet, A, Le Gorrec, M, Machillot, P, Neveu, B, Thielens, N, Malissen, B, Bonneville, M, Housset, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope.
J.Immunol., 183, 2009
|
|
4ISB
 
 | |
4IV6
 
 | |
2QP2
 
 | | Structure of a MACPF/perforin-like protein | | Descriptor: | CALCIUM ION, Unknown protein | | Authors: | Rosado, C.J, Buckle, A.M, Law, R.H.P, Butcher, R.E, Kan, W.T, Bird, C.H, Ung, K, Browne, K.A, Baran, K, Bashtannyk-Puhalovich, T.A, Faux, N.G, Wong, W, Porter, C.J, Pike, R.N, Ellisdon, A.M, Pearce, M.C, Bottomley, S.P, Emsley, J, Smith, A.I, Rossjohn, J, Hartland, E.L, Voskoboinik, I, Trapani, J.A, Bird, P.I, Dunstone, M.A, Whisstock, J.C. | | Deposit date: | 2007-07-22 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A common fold mediates vertebrate defense and bacterial attack
Science, 317, 2007
|
|
