4DB1
 
 | | Cardiac human myosin S1dC, beta isoform complexed with Mn-AMPPNP | | Descriptor: | MANGANESE (II) ION, Myosin-7, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Klenchin, V.A, Deacon, J.C, Combs, A.C, Leinwand, L.A, Rayment, I. | | Deposit date: | 2012-01-13 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cardiac human myosin S1dC, beta isoform complexed with Mn-AMPPNP
To be Published
|
|
4DER
 
 | |
2HY7
 
 | |
2O4T
 
 | |
2HYY
 
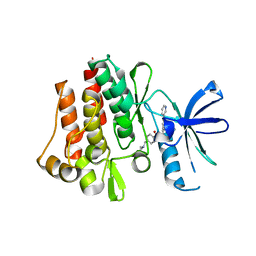 | | Human Abl kinase domain in complex with imatinib (STI571, Glivec) | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Liebetanz, J, Fabbro, D, Manley, P. | | Deposit date: | 2006-08-08 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia.
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
4DGA
 
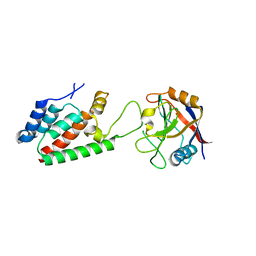 | | TRIMCyp cyclophilin domain from Macaca mulatta: HIV-1 CA(O-loop) complex | | Descriptor: | TRIMCyp, capsid protein | | Authors: | Caines, M.E.C, Bichel, K, Price, A.J, McEwan, W.A, James, L.C. | | Deposit date: | 2012-01-25 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Diverse HIV viruses are targeted by a conformationally dynamic antiviral.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3ODP
 
 | |
4DGN
 
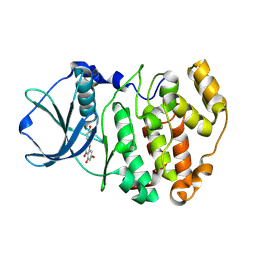 | | Crystal Structure of maize CK2 in complex with the inhibitor luteolin | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, Casein kinase II subunit alpha | | Authors: | Lolli, G, Mazzorana, M, Battistutta, R. | | Deposit date: | 2012-01-26 | | Release date: | 2012-08-01 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inhibition of protein kinase CK2 by flavonoids and tyrphostins. A structural insight.
Biochemistry, 51, 2012
|
|
3OE1
 
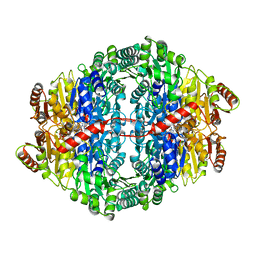 | | Pyruvate decarboxylase variant Glu473Asp from Z. mobilis in complex with reaction intermediate 2-lactyl-ThDP | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Meyer, D, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Double duty for a conserved glutamate in pyruvate decarboxylase: evidence of the participation in stereoelectronically controlled decarboxylation and in protonation of the nascent carbanion/enamine intermediate .
Biochemistry, 49, 2010
|
|
2OA0
 
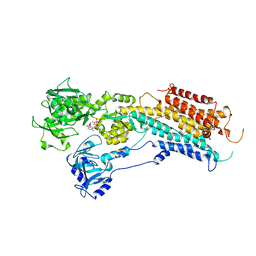 | | Crystal structure of Calcium ATPase with bound ADP and cyclopiazonic acid | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Young, H.S, Moncoq, K.A. | | Deposit date: | 2006-12-14 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The molecular basis for cyclopiazonic Acid inhibition of the sarcoplasmic reticulum calcium pump.
J.Biol.Chem., 282, 2007
|
|
3OF8
 
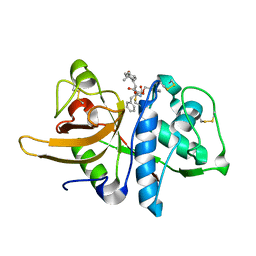 | |
3OFE
 
 | |
4DAL
 
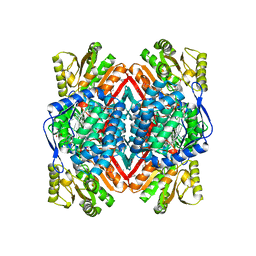 | | Crystal structure of Putative aldehyde dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | GLYCEROL, Putative aldehyde dehydrogenase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-01-12 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Putative aldehyde dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
3OFG
 
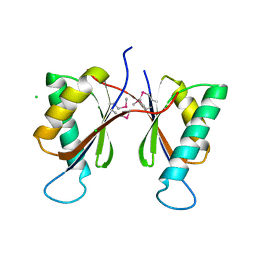 | | Structured Domain of Caenorhabditis elegans BMY-1 | | Descriptor: | Boca/mesd chaperone for ywtd beta-propeller-egf protein 1, CHLORIDE ION | | Authors: | Collins, M.N, Hendrickson, W.A. | | Deposit date: | 2010-08-14 | | Release date: | 2011-03-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.367 Å) | | Cite: | Structural Characterization of the Boca/Mesd Maturation Factors for LDL-Receptor-Type beta-Propeller Domains
Structure, 2011
|
|
3OG6
 
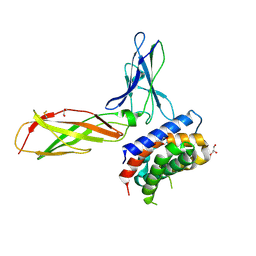 | | The crystal structure of human interferon lambda 1 complexed with its high affinity receptor in space group P212121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Interleukin 28 receptor, ... | | Authors: | Miknis, Z.J, Magracheva, E, Lei, W, Zdanov, A, Kotenko, S.V, Wlodawer, A. | | Deposit date: | 2010-08-16 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of the complex of human interferon-lambda1 with its high affinity receptor interferon-lambdaR1.
J.Mol.Biol., 404, 2010
|
|
3OGC
 
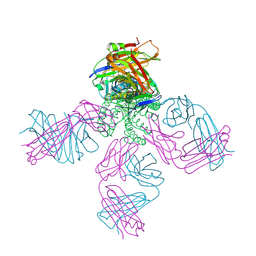 | | KcsA E71A variant in presence of Na+ | | Descriptor: | SODIUM ION, Voltage-gated potassium channel, antibody Fab fragment heavy chain, ... | | Authors: | McCoy, J.G, Nimigean, C.M. | | Deposit date: | 2010-08-16 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Mechanism for selectivity-inactivation coupling in KcsA potassium channels.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4DDY
 
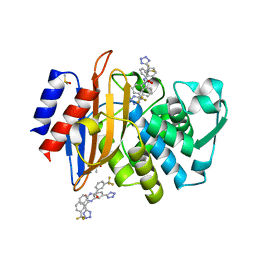 | | CTX-M-9 class A beta-lactamase complexed with compound 10 | | Descriptor: | Beta-lactamase, DIMETHYL SULFOXIDE, N-[3-(1H-tetrazol-5-yl)phenyl]-3-(trifluoromethyl)benzamide | | Authors: | Nichols, D.A, Chen, Y. | | Deposit date: | 2012-01-19 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure-Based Design of Potent and Ligand-Efficient Inhibitors of CTX-M Class A Beta-Lactamase
J.Med.Chem., 55, 2012
|
|
4DQ1
 
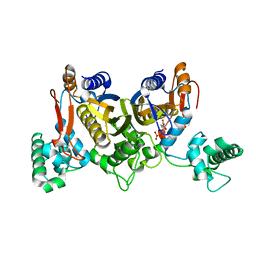 | | Thymidylate synthase from Staphylococcus aureus. | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Osipiuk, J, Holowicki, J, Jedrzejczak, R, Rubin, E, Guinn, K, Ioerger, T, Baker, D, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-02-14 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Thymidylate synthase from Staphylococcus aureus.
To be Published
|
|
3OBZ
 
 | |
4DQE
 
 | | Crystal Structure of (G16C/L38C) HIV-1 Protease in Complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, Aspartyl protease | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-02-15 | | Release date: | 2012-03-07 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease.
J.Am.Chem.Soc., 134, 2012
|
|
2OIP
 
 | |
2NSC
 
 | |
2OD7
 
 | | Crystal Structure of yHst2 bound to the intermediate analogue ADP-HPD, and and aceylated H4 peptide | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, Acetylated histone H4 peptide, NAD-dependent deacetylase HST2, ... | | Authors: | Marmorstein, R.Q, Sanders, B.D. | | Deposit date: | 2006-12-21 | | Release date: | 2007-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for nicotinamide inhibition and base exchange in sir2 enzymes.
Mol.Cell, 25, 2007
|
|
2NVH
 
 | |
2OEW
 
 | | Structure of ALIX/AIP1 Bro1 Domain | | Descriptor: | Programmed cell death 6-interacting protein | | Authors: | Fisher, R.D, Zhai, Q, Robinson, H, Hill, C.P. | | Deposit date: | 2007-01-01 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Biochemical Studies of ALIX/AIP1 and Its Role in Retrovirus Budding
Cell(Cambridge,Mass.), 128, 2007
|
|
