1RI3
 
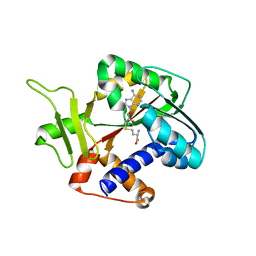 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
8SXP
 
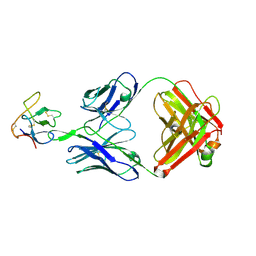 | |
1RO8
 
 | | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analogue, cytidine-5'-monophosphate | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, alpha-2,3/8-sialyltransferase | | Authors: | Chiu, C.P, Watts, A.G, Lairson, L.L, Gilbert, M, Lim, D, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2003-12-01 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog.
Nat.Struct.Mol.Biol., 11, 2004
|
|
8SXE
 
 | |
1RJX
 
 | | Human PLASMINOGEN CATALYTIC DOMAIN, K698M MUTANT | | Descriptor: | Plasminogen, SULFATE ION | | Authors: | Terzyan, S, Wakeham, N, Zhai, P, Rodgers, K, Zhang, X.C. | | Deposit date: | 2003-11-20 | | Release date: | 2003-12-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Lys-698 to met substitution in human plasminogen catalytic domain
Proteins, 56, 2004
|
|
1RPO
 
 | |
1ROP
 
 | |
1RQS
 
 | | NMR structure of C-terminal domain of ribosomal protein L7 from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
8SVG
 
 | | Ubiquitin variant i53 in complex with 53BP1 Tudor domain | | Descriptor: | Tumor protein p53 binding protein 1, Ubiquitin variant i53 | | Authors: | Holden, J.K, Partridge, J.R, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
1RK5
 
 | | The D-aminoacylase mutant D366A in complex with 100mM CuCl2 | | Descriptor: | ACETATE ION, COPPER (II) ION, D-aminoacylase, ... | | Authors: | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
J.Biol.Chem., 279, 2004
|
|
1RSV
 
 | | azide complex of the diferrous E238A mutant R2 subunit of ribonucleotide reductase | | Descriptor: | AZIDE ION, FE (III) ION, MERCURY (II) ION, ... | | Authors: | Assarsson, M, Andersson, M.E, Hogbom, M, Persson, B.O, Sahlin, M, Barra, A.L, Sjoberg, B.M, Nordlund, P, Graslund, A. | | Deposit date: | 2003-12-10 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Restoring proper radical generation by azide binding to the iron site of the E238A mutant R2 protein of ribonucleotide reductase from Escherichia coli.
J.Biol.Chem., 276, 2001
|
|
8SVH
 
 | | Ubiquitin variant i53 mutant L67R bound to 53BP1 Tudor Domain | | Descriptor: | Tumor protein p53 binding protein 1, Ubiquitin variant i53: mutant L67R | | Authors: | Holden, J.K, Partridge, J.R, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
1RTE
 
 | | X-ray Structure of Cyanide Derivative of Truncated Hemoglobin N (trHbN) from Mycobacterium Tuberculosis | | Descriptor: | CYANIDE ION, Hemoglobin-like protein HbN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Milani, M, Guertin, M, Boffi, A, Antonini, G, Bocedi, A, Mattu, M, Bolognesi, M, Ascenzi, P. | | Deposit date: | 2003-12-10 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cyanide binding to truncated hemoglobins: a crystallographic and kinetic study
Biochemistry, 43, 2004
|
|
1R1F
 
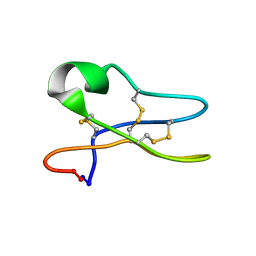 | | Solution Structure of the Cyclotide Palicourein: Implications for the development of pharmaceutical and agricultural applications | | Descriptor: | Palicourein | | Authors: | Barry, D.G, Daly, N.L, Bokesch, H.R, Gustafson, K.R, Craik, D.J. | | Deposit date: | 2003-09-23 | | Release date: | 2004-04-06 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cyclotide palicourein: implications for the development of a pharmaceutical framework.
STRUCTURE, 12, 2004
|
|
8SNK
 
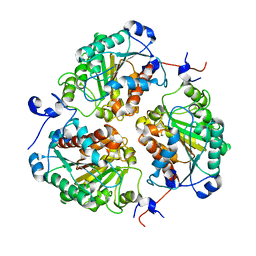 | | Crystal structure of metformin hydrolase (MfmAB) from Pseudomonas mendocina sp. MET-2 mutant (MfmA/D188N) | | Descriptor: | ZINC ION, metformin hydrolase subunit A, metformin hydrolase subunit B | | Authors: | Tassoulas, L.J, Rankin, J.A, Elias, M.H, Wackett, L.P. | | Deposit date: | 2023-04-27 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dinickel enzyme evolved to metabolize the pharmaceutical metformin and its implications for wastewater and human microbiomes.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8S96
 
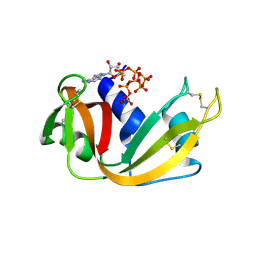 | | RNase A-Adenosine 5'-Heptaphosphate (RNaseA.p7A) | | Descriptor: | Ribonuclease pancreatic, adenosine 5'-heptaphosphate | | Authors: | Park, G, Cummins, C. | | Deposit date: | 2023-03-27 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Pentaphosphorylation via the Anhydride of Dihydrogen Pentametaphosphate: Access to Nucleoside Hexa- and Heptaphosphates and Study of Their Interaction with Ribonuclease A.
Acs Cent.Sci., 10, 2024
|
|
8SVI
 
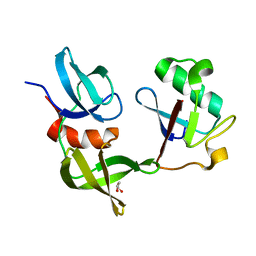 | | Ubiquitin variant i53:Mutant L67H with 53BP1 Tudor domain | | Descriptor: | GLYCEROL, Tumor protein p53 binding protein 1, Ubiquitin Variant i53: Mutant L67H | | Authors: | Partridge, J.R, Holden, J.K, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
8SMK
 
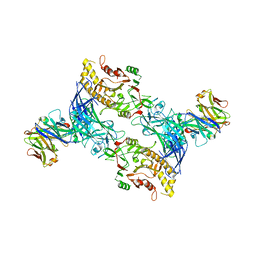 | | hPAD4 bound to Activating Fab hA362 | | Descriptor: | Activating Fab 362 heavy chain, Activating Fab 362 light chain, CALCIUM ION, ... | | Authors: | Maker, A, Verba, K.A. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Antibody discovery identifies regulatory mechanisms of protein arginine deiminase 4.
Nat.Chem.Biol., 20, 2024
|
|
8S9Z
 
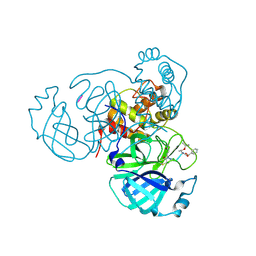 | | Mpro inhibitors of SARS-CoV-2 | | Descriptor: | 3C-like proteinase nsp5, Mpro inhibitor | | Authors: | Blankenship, L.R, Liu, W.R. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of MPI89 with Mpro of SARS-CoV-2 at 1.85A resolution.
To Be Published
|
|
8SVJ
 
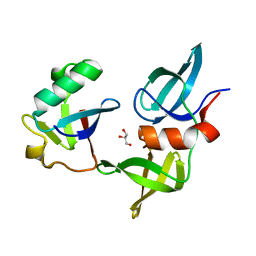 | | Ubiquitin variant i53: mutant VHH with 53BP1 Tudor domain | | Descriptor: | GLYCEROL, Tumor protein p53 binding protein 1, Ubiquitin varient i53 mutant VHH | | Authors: | Holden, J, Partridge, J.R, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
1R3R
 
 | |
1R45
 
 | | ADP-ribosyltransferase C3bot2 from Clostridium botulinum, triclinic form | | Descriptor: | GLYCEROL, Mono-ADP-ribosyltransferase C3, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L, Narumiya, S. | | Deposit date: | 2003-10-03 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of ADP-ribosyltransferase C3bot2 from Clostridium botulinum
To be Published
|
|
8T0O
 
 | | Fab from mAb RB2AT_87 | | Descriptor: | CHLORIDE ION, RB2AT_87 Fab Heavy chain, RB2AT_87 Fab Light chain | | Authors: | Kreutzer, A.G, Malonis, R.J, Lai, J.R, Nowick, J.S. | | Deposit date: | 2023-06-01 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Generation and Study of Antibodies against Two Triangular Trimers Derived from A beta.
Pept Sci (Hoboken), 116, 2024
|
|
1R50
 
 | | Bacillus subtilis lipase A with covalently bound Sc-IPG-phosphonate-inhibitor | | Descriptor: | Lipase, [(4S)-2,2-DIMETHYL-1,3-DIOXOLAN-4-YL]METHYL HYDROGEN HEX-5-ENYLPHOSPHONATE | | Authors: | Droege, M.J, Van Pouderoyen, G, Vrenken, T.E, Rueggeberg, C.J, Reetz, M.T, Dijkstra, B.W, Quax, W.J. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Directed Evolution of Bacillus subtilis Lipase A by Use of Enantiomeric Phosphonate Inhibitors: Crystal Structures and Phage Display Selection
Chembiochem, 7, 2005
|
|
8SML
 
 | | hPAD4 bound to inhibitory Fab hI365 | | Descriptor: | CALCIUM ION, Fab hI365 heavy chain, Fab hI365 light chain, ... | | Authors: | Maker, A, Verba, K.A. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Antibody discovery identifies regulatory mechanisms of protein arginine deiminase 4.
Nat.Chem.Biol., 20, 2024
|
|
