5G0N
 
 | | Structure of rat neuronal nitric oxide synthase D597N mutant heme domain in complex with N1-(5-(2-(6-AMINO-4-METHYLPYRIDIN-2-YL)ETHYL) PYRIDIN-3-YL)-N1,N2-DIMETHYLETHANE-1,2-DIAMINE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N1-(5-(2-(6-Amino-4-methylpyridin-2-yl)ethyl)pyridin-3-yl)-N1,N2-dimethylethane-1,2-diamine, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2016-03-21 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | Electrostatic Control of Isoform Selective Inhibitor Binding in Nitric Oxide Synthase.
Biochemistry, 55, 2016
|
|
4QQU
 
 | | Crystal structure of the cobalamin-independent methionine synthase enzyme in a closed conformation | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, N-[4-({[(6S)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-gamma-glutamyl-L-gamma-glutamyl-L-glutamic acid, ... | | Authors: | Ubhi, D.K, Robertus, J.D. | | Deposit date: | 2014-06-29 | | Release date: | 2015-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The cobalamin-independent methionine synthase enzyme captured in a substrate-induced closed conformation.
J.Mol.Biol., 427, 2015
|
|
7P33
 
 | | Epstein-Barr virus encoded Bcl-2 homolog BHRF-1 in complex with Bid BH3 peptide | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BHRF1, BH3-interacting domain death agonist p15, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2021-07-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.78542733 Å) | | Cite: | Crystal Structures of Epstein-Barr Virus Bcl-2 Homolog BHRF1 Bound to Bid and Puma BH3 Motif Peptides.
Viruses, 14, 2022
|
|
3LA1
 
 | |
5MBV
 
 | | Cryo-EM structure of Lambda Phage protein GamS bound to RecBCD. | | Descriptor: | Host-nuclease inhibitor protein gam, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Wilkinson, M, Chaban, Y, Wigley, D.B. | | Deposit date: | 2016-11-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the inhibition of RecBCD by Gam and its synergistic antibacterial effect with quinolones.
Elife, 5, 2016
|
|
5AVW
 
 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 16.5 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5FIL
 
 | | Bovine mitochondrial ATP synthase state 3b | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
5ME6
 
 | | Crystal Structure of eiF4E from C. melo bound to a CAP analog | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic transcription initiation factor 4E | | Authors: | Querol-Audi, J, Silva, C, Miras, M, Aranda-Regules, M, Verdaguer, N. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of eIF4E in Complex with an eIF4G Peptide Supports a Universal Bipartite Binding Mode for Protein Translation.
Plant Physiol., 174, 2017
|
|
3LB9
 
 | | Crystal structure of the B. circulans cpA123 circular permutant | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | D'Angelo, I, Reitinger, S, Ludwiczek, M, Strynadka, N, Withers, S.G, Mcintosh, L.P. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Circular permutation of Bacillus circulans xylanase: a kinetic and structural study.
Biochemistry, 49, 2010
|
|
5AW8
 
 | | Kinetics by X-ray crystallography: E2.MgF42-.2RB+ crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5FLM
 
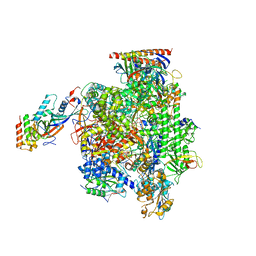 | | Structure of transcribing mammalian RNA polymerase II | | Descriptor: | DNA, DNA-RNA ELONGATION SCAFFOLD, DNA-DIRECTED RNA POLYMERASE, ... | | Authors: | Bernecky, C, Herzog, F, Baumeister, W, Plitzko, J.M, Cramer, P. | | Deposit date: | 2015-10-26 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of Transcribing Mammalian RNA Polymerase II
Nature, 529, 2016
|
|
5MMM
 
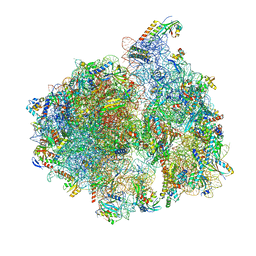 | | Structure of the 70S chloroplast ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein 2, ... | | Authors: | Bieri, P, Leibundgut, M, Saurer, M, Boehringer, D, Ban, N. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The complete structure of the chloroplast 70S ribosome in complex with translation factor pY.
EMBO J., 36, 2017
|
|
5FBA
 
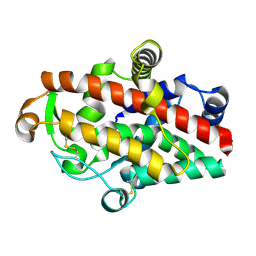 | | S1 nuclease from Aspergillus oryzae in complex with phosphate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Nuclease S1, ... | | Authors: | Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Catalytic Properties of S1 Nuclease from Aspergillus oryzae Responsible for Substrate Recognition, Cleavage, Non-Specificity, and Inhibition.
PLoS ONE, 11, 2016
|
|
6XAT
 
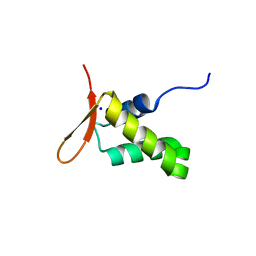 | | Crystal structure of the human FoxP4 DNA binding Domain | | Descriptor: | FOXP4 protein, SODIUM ION | | Authors: | VIllalobos, P, Castro-Fernandez, V, Medina, E, Gonzalez-Ordenes, F, Maturana, P, Herrera-Morande, A, Ramirez-Sarmiento, C.A, Babul, J. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unraveling the folding and dimerization properties of the human FoxP subfamily of transcription factors.
Febs Lett., 597, 2023
|
|
5FBD
 
 | | S1 nuclease from Aspergillus oryzae in complex with phosphate and 2'-deoxycytidine | | Descriptor: | 2'-DEOXYCYTIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nuclease S1, ... | | Authors: | Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Catalytic Properties of S1 Nuclease from Aspergillus oryzae Responsible for Substrate Recognition, Cleavage, Non-Specificity, and Inhibition.
PLoS ONE, 11, 2016
|
|
5AWW
 
 | | Precise Resting State of Thermus thermophilus SecYEG | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Protein translocase subunit SecE, Protein translocase subunit SecY, ... | | Authors: | Tanaka, Y, Sugano, Y, Takemoto, M, Kusakizako, T, Kumazaki, K, Ishitani, R, Nureki, O, Tsukazaki, T. | | Deposit date: | 2015-07-10 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.724 Å) | | Cite: | Crystal Structures of SecYEG in Lipidic Cubic Phase Elucidate a Precise Resting and a Peptide-Bound State.
Cell Rep, 13, 2015
|
|
5FBJ
 
 | | Complex structure of JMJD5 and substrate | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Lysine-specific demethylase 8, ... | | Authors: | Liu, H.L, Wang, Y, Wang, C, Zhang, G.Y. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | to be published
To Be Published
|
|
5FBZ
 
 | | Structure of subtilase SubHal from Bacillus halmapalus - complex with chymotrypsin inhibitor CI2A | | Descriptor: | Autoproteolytic fragment of enzyme subtilase SubHal, CALCIUM ION, Enzyme subtilase SubHal from Bacillus halmapalus, ... | | Authors: | Dohnalek, J, Brzozowski, A.M, Svendsen, A, Wilson, K.S. | | Deposit date: | 2015-12-14 | | Release date: | 2016-05-18 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Stabilization of Enzymes by Metal Binding: Structures of Two Alkalophilic Bacillus Subtilases and Analysis of the Second Metal-Binding Site of the Subtilase Family
Book, 2016
|
|
6XBO
 
 | |
5FCG
 
 | |
5B34
 
 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles in Complex with Iodine-labeled Detergent HAD13a Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | 2,4,6-tris(iodanyl)-5-(octanoylamino)benzene-1,3-dicarboxylic acid, Bacteriorhodopsin, DECANE, ... | | Authors: | Mizohata, E, Nakane, T. | | Deposit date: | 2016-02-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5M7O
 
 | | Crystal structure of NtrX from Brucella abortus processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | MAGNESIUM ION, Nitrogen assimilation regulatory protein | | Authors: | Cornaciu, I, Fernandez, I, Hoffmann, G, Carrica, M.C, Goldbaum, F.A, Marquez, J.A. | | Deposit date: | 2016-10-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-Dimensional Structure of Full-Length NtrX, an Unusual Member of the NtrC Family of Response Regulators.
J. Mol. Biol., 429, 2017
|
|
6XCX
 
 | |
5M7W
 
 | | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSpG mRNA 5' cap analog (beta-S-ARCA D2) | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(sulfanyl)phosphoryl] hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSpG mRNA 5' cap analog (beta-S-ARCA D2)
To Be Published
|
|
5J4Z
 
 | | Architecture of tight respirasome | | Descriptor: | COMPLEX I 13KDA/NDUFS6, COMPLEX I 15KDA/NDUFS5, COMPLEX I 18KDA/NDUFS6, ... | | Authors: | Letts, J.A, Fiedorczuk, K, Sazanov, L.A. | | Deposit date: | 2016-04-01 | | Release date: | 2016-09-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | The architecture of respiratory supercomplexes.
Nature, 537, 2016
|
|
