7E6E
 
 | | Crystal structure of PMP-bound form of cysteine desulfurase SufS R376A from Bacillus subtilis in D-cycloserine-inhibition | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6A
 
 | | Crystal structure of cysteine desulfurase SufS C361A from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6C
 
 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase SufS C361A from Bacillus subtilis | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, 1,2-ETHANEDIOL, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6D
 
 | |
7E6B
 
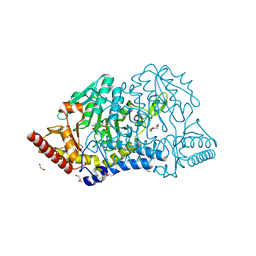 | | Crystal structure of PMP-bound form of cysteine desulfurase SufS C361A from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6F
 
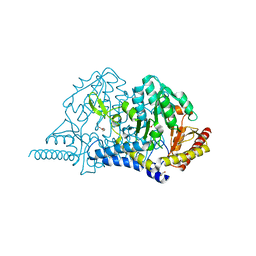 | | Crystal structure of PMP-bound form of cysteine desulfurase SufS R376A from Bacillus subtilis in L-cycloserine-inhibition | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
5MPR
 
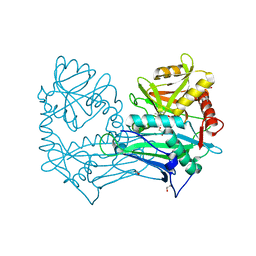 | | Single Amino Acid Variant of Human Mitochondrial Branched Chain Amino Acid Aminotransferase 2 | | Descriptor: | 1,2-ETHANEDIOL, Branched-chain-amino-acid aminotransferase, mitochondrial, ... | | Authors: | Hakansson, M, Walse, B, Nilsson, C, Anderson, L.C. | | Deposit date: | 2016-12-18 | | Release date: | 2017-07-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Intact Protein Analysis at 21 Tesla and X-Ray Crystallography Define Structural Differences in Single Amino Acid Variants of Human Mitochondrial Branched-Chain Amino Acid Aminotransferase 2 (BCAT2).
J. Am. Soc. Mass Spectrom., 28, 2017
|
|
1MM0
 
 | | Solution structure of termicin, an antimicrobial peptide from the termite Pseudacanthotermes spiniger | | Descriptor: | Termicin | | Authors: | Da Silva, P, Jouvensal, L, Lamberty, M, Bulet, P, Caille, A, Vovelle, F. | | Deposit date: | 2002-09-02 | | Release date: | 2003-05-13 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of termicin, an antimicrobial peptide from the termite Pseudacanthotermes spiniger
PROTEIN SCI., 12, 2003
|
|
6BCA
 
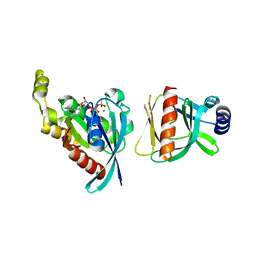 | |
5BYY
 
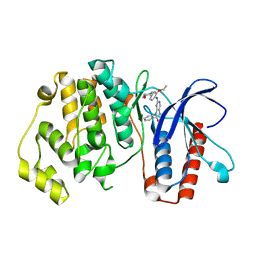 | | ERK5 IN COMPLEX WITH SMALL MOLECULE | | Descriptor: | 2-{[2-ethoxy-4-(4-hydroxypiperidin-1-yl)phenyl]amino}-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Mitogen-activated protein kinase 7 | | Authors: | Chen, H, Tucker, J, Wang, X, Gavine, P.R, Philips, C, Augustin, M.A, Schreiner, P, Steinbacher, S, Preston, M, Ogg, D. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
9O38
 
 | | Transmembrane domains of the human sweet receptor (TAS1R2 + TAS1R3) from Class 3 particles (rigidly fitted from PDB:9NOX and 9NOR) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Nanobody 35 (NB35), ... | | Authors: | Juen, Z, Lu, Z, Yu, R, Chang, A.N, Wang, B, Fitzpatrick, A.W.P, Zuker, C.S. | | Deposit date: | 2025-04-06 | | Release date: | 2025-05-14 | | Last modified: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The structure of human sweetness.
Cell, 188, 2025
|
|
1J3G
 
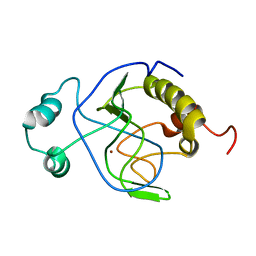 | | Solution structure of Citrobacter Freundii AmpD | | Descriptor: | AmpD protein, ZINC ION | | Authors: | Liepinsh, E, Genereux, C, Dehareng, D, Joris, B, Otting, G. | | Deposit date: | 2003-01-31 | | Release date: | 2003-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Citrobacter freundii AmpD, Comparison with Bacteriophage T7 Lysozyme
and Homology with PGRP Domains
J.Mol.Biol., 327, 2003
|
|
5N5H
 
 | | Crystal structure of metallo-beta-lactamase VIM-1 in complex with ML302F inhibitor | | Descriptor: | (2Z)-2-sulfanyl-3-(2,3,6-trichlorophenyl)prop-2-enoic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Salimraj, R, Hinchliffe, P, Spencer, J. | | Deposit date: | 2017-02-14 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of VIM-1 complexes explain active site heterogeneity in VIM-class metallo-beta-lactamases.
FEBS J., 286, 2019
|
|
7FEV
 
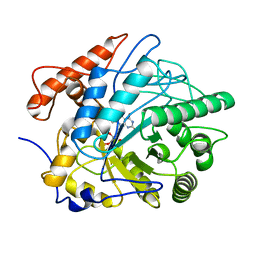 | | Crystal structure of Old Yellow Enzyme6 (OYE6) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN binding | | Authors: | Singh, Y, Sharma, R, Mishra, M, Verma, P.K, Saxena, A.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Crystal structure of ArOYE6 reveals a novel C-terminal helical extension and mechanistic insights into the distinct class III OYEs from pathogenic fungi.
Febs J., 289, 2022
|
|
1MZT
 
 | |
5N5I
 
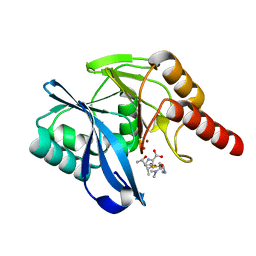 | | Crystal Structure of VIM-1 metallo-beta-lactamase in complex with hydrolysed meropenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Salimraj, R, Hinchliffe, P, Spencer, J. | | Deposit date: | 2017-02-14 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of VIM-1 complexes explain active site heterogeneity in VIM-class metallo-beta-lactamases.
FEBS J., 286, 2019
|
|
5N5G
 
 | |
6H86
 
 | |
6H36
 
 | | The crystal structure of human carbonic anhydrase VII in complex with 4-(4-phenylpiperidine-1-carbonyl)benzenesulfonamide. | | Descriptor: | 4-(4-phenylpiperidin-1-yl)carbonylbenzenesulfonamide, Carbonic anhydrase 7, ZINC ION | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
6H38
 
 | | The crystal structure of human carbonic anhydrase VII in complex with 4-[(4-fluorophenyl)methyl]-1-piperazinyl]benzenesulfonamide. | | Descriptor: | 4-[4-[(4-fluorophenyl)methyl]piperazin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 7, GLYCEROL, ... | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
1VHV
 
 | | Crystal structure of diphthine synthase | | Descriptor: | diphthine synthase | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VIC
 
 | | Crystal structure of CMP-KDO synthetase | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VIV
 
 | | Crystal structure of a hypothetical protein | | Descriptor: | Hypothetical protein yckF | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VGW
 
 | |
1VGX
 
 | |
