8H1W
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase | | Authors: | Takahashi, A, Murakami, T, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H1Q
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-serine | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-03 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
7O58
 
 | | Human phosphomannomutase 2 (PMM2) with mutation T237M in complex with the activator glucose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ramon-Maiques, S, Briso-Montiano, A, Del Cano-Ochoa, F, Vilas, A, Perez, B, Rubio, V. | | Deposit date: | 2021-04-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Insight on molecular pathogenesis and pharmacochaperoning potential in phosphomannomutase 2 deficiency, provided by novel human phosphomannomutase 2 structures.
J Inherit Metab Dis, 45, 2022
|
|
7O4G
 
 | | Human phosphomannomutase 2 (PMM2) wild-type soaked with the activator glucose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ramon-Maiques, S, Briso-Montiano, A, Del Cano-Ochoa, F, Vilas, A, Perez, B, Rubio, V. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Insight on molecular pathogenesis and pharmacochaperoning potential in phosphomannomutase 2 deficiency, provided by novel human phosphomannomutase 2 structures.
J Inherit Metab Dis, 45, 2022
|
|
8QV9
 
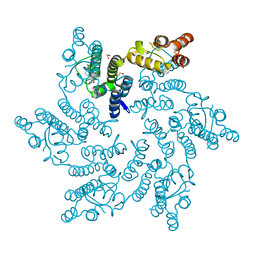 | | Hexameric HIV-1 CA in complex with DDD01829021 | | Descriptor: | 1,2-ETHANEDIOL, 7-bromanyl-3-(phenylmethyl)-1~{H}-benzimidazol-2-one, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
6VKE
 
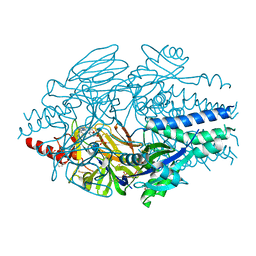 | | Crystal Structure of Inhibitor JNJ-40012665 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-(5-chloro-2-{[1-(3,4-dimethoxyphenyl)-2-oxo-1,2-dihydro-3H-imidazo[4,5-c]pyridin-3-yl]methyl}-1H-indol-1-yl)butanenitrile, CHLORIDE ION, ... | | Authors: | McLellan, J.S. | | Deposit date: | 2020-01-20 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of 3-({5-Chloro-1-[3-(methylsulfonyl)propyl]-1H-indol-2-yl}methyl)-1-(2,2,2-trifluoroethyl)-1,3-dihydro-2H-imidazo[4,5-c]pyridin-2-one (JNJ-53718678), a Potent and Orally Bioavailable Fusion Inhibitor of Respiratory Syncytial Virus.
J.Med.Chem., 63, 2020
|
|
6VKV
 
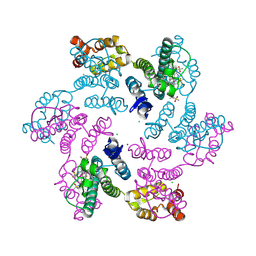 | |
8QD3
 
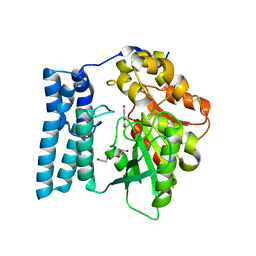 | | Ayg1p in complex with 1,3,6,8-Tetrahydroxynaphthalene | | Descriptor: | 1,2-ETHANEDIOL, FLAVIOLIN, Pigment biosynthesis protein yellowish-green 1, ... | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
7R73
 
 | |
6XQI
 
 | | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A | | Descriptor: | ASN-PRO-LEU-GLU-PHE-LEU, Protein Vpr, UV excision repair protein RAD23 homolog A, ... | | Authors: | Calero, G.C, Wu, Y, Weiss, S.C. | | Deposit date: | 2020-07-09 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A.
Nat Commun, 12, 2021
|
|
7LOH
 
 | | Structure of the HIV-1 gp41 transmembrane domain and cytoplasmic tail | | Descriptor: | Transmembrane protein gp41 | | Authors: | Piai, A, Fu, Q, Sharp, A.K, Bighi, B, Brown, A.M, Chou, J.J. | | Deposit date: | 2021-02-10 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Model of the Entire Membrane-Interacting Region of the HIV-1 Fusion Protein and Its Perturbation of Membrane Morphology.
J.Am.Chem.Soc., 143, 2021
|
|
8E4H
 
 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAGAGGAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*AP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*TP*CP*TP*TP*AP*T)-3'), Transcription factor PU.1 | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8E3R
 
 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAAAGGAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*AP*AP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*TP*TP*TP*TP*AP*T)-3'), Transcription factor PU.1 | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-08-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8EK3
 
 | |
8EKV
 
 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAGCGGATGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*CP*GP*GP*AP*TP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*AP*TP*CP*CP*GP*CP*TP*TP*AP*T)-3'), SODIUM ION, ... | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-09-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8ENG
 
 | |
8E3K
 
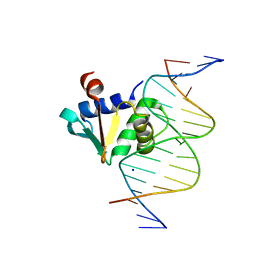 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAGCGGAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*CP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*GP*CP*TP*TP*AP*T)-3'), SODIUM ION, ... | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-08-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
6Y23
 
 | |
8EKZ
 
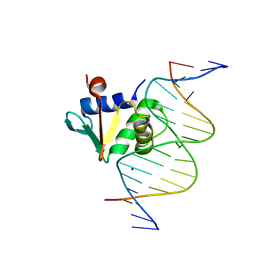 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAGGAGAAGTAGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*GP*AP*GP*AP*AP*GP*TP*AP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*AP*CP*TP*TP*CP*TP*CP*CP*TP*TP*AP*T)-3'), SODIUM ION, ... | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-09-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8EMD
 
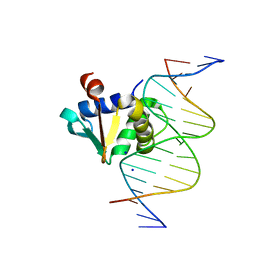 | |
8E5Y
 
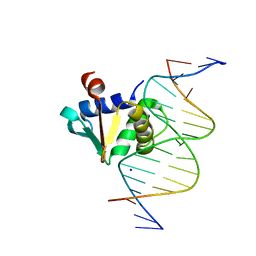 | |
8EE9
 
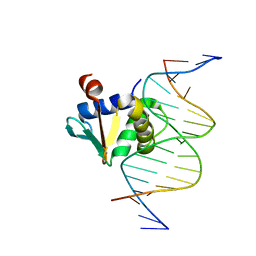 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAAAGGAATGGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*AP*AP*GP*GP*AP*AP*TP*GP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*CP*AP*TP*TP*CP*CP*TP*TP*TP*TP*AP*T)-3'), Transcription factor PU.1 | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-09-06 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
6GPJ
 
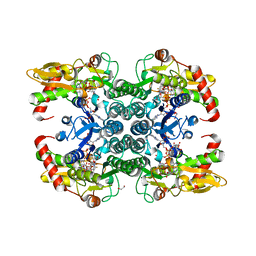 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase in complex with GDP-4F-Man | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, GDP-mannose 4,6 dehydratase, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
8EK8
 
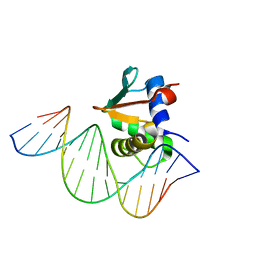 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAAAGGAGAAGGG) | | Descriptor: | DNA (5'-D(*AP*TP*AP*AP*AP*AP*GP*GP*AP*GP*AP*AP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*TP*TP*CP*TP*CP*CP*TP*TP*TP*TP*AP*T)-3'), Transcription factor PU.1 | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-09-20 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
9C7L
 
 | | Crystal structure of pentalenene synthase variant F76A complexed with 2-fluorofarnesyl diphosphate | | Descriptor: | (2Z,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, MAGNESIUM ION, Pentalenene synthase | | Authors: | Prem Kumar, R, Ellenburg, W.H, Oprian, D.D. | | Deposit date: | 2024-06-10 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Caryolan-1-ol Synthase, a Sesquiterpene Synthase Catalyzing an Initial Anti-Markovnikov Cyclization Reaction.
Biochemistry, 63, 2024
|
|
