1PDX
 
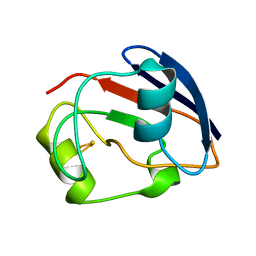 | | PUTIDAREDOXIN | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, PROTEIN (PUTIDAREDOXIN) | | Authors: | Pochapsky, T.C, Jain, N.U, Kuti, M, Lyons, T.A, Heymont, J. | | Deposit date: | 1999-02-15 | | Release date: | 1999-05-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A refined model for the solution structure of oxidized putidaredoxin.
Biochemistry, 38, 1999
|
|
1B9Q
 
 | |
1B9S
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN THE ACTIVE SITE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(N-ACETYLAMINO)-3-[N-(2-ETHYLBUTANOYLAMINO)]BENZOIC ACID, CALCIUM ION, ... | | Authors: | Finley, J.B, Atigadda, V.R, Duarte, F, Zhao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 1999-02-15 | | Release date: | 1999-02-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
1B9V
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN TEH ACTIVE SITE | | Descriptor: | 1-[4-CARBOXY-2-(3-PENTYLAMINO)PHENYL]-5,5'-DI(HYDROXYMETHYL)PYRROLIDIN-2-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Finley, J.B, Atigadda, V.R, Duarte, F, Zahao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 1999-02-15 | | Release date: | 1999-02-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
1B9W
 
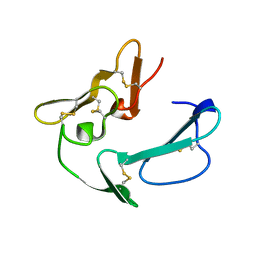 | | C-TERMINAL MEROZOITE SURFACE PROTEIN 1 FROM PLASMODIUM CYNOMOLGI | | Descriptor: | PROTEIN (MEROZOITE SURFACE PROTEIN 1) | | Authors: | Bentley, G.A, Chitarra, V, Holm, I, Longacre, S. | | Deposit date: | 1999-02-15 | | Release date: | 1999-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of C-terminal merozoite surface protein 1 at 1.8 A resolution, a highly protective malaria vaccine candidate.
Mol.Cell, 3, 1999
|
|
1B9R
 
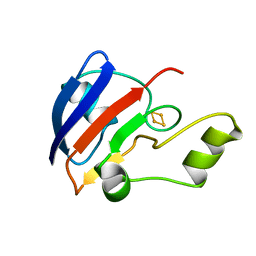 | | TERPREDOXIN FROM PSEUDOMONAS SP. | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, PROTEIN (TERPREDOXIN) | | Authors: | Mo, H, Pochapsky, S.S, Pochapsky, T.C. | | Deposit date: | 1999-02-15 | | Release date: | 1999-05-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A model for the solution structure of oxidized terpredoxin, a Fe2S2 ferredoxin from Pseudomonas.
Biochemistry, 38, 1999
|
|
1B9P
 
 | |
1B9T
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN THE ACTIVE SITE | | Descriptor: | 1-(4-CARBOXY-2-GUANIDINOPENTYL)-5,5'-DI(HYDROXYMETHYL)PYRROLIDIN-2-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Finley, J.B, Atigadda, V.R, Duarte, F, Zhao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 1999-02-15 | | Release date: | 1999-02-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
1B9U
 
 | |
2SFP
 
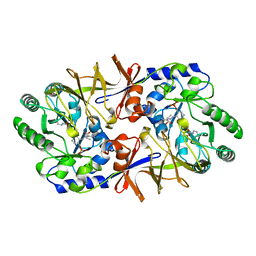 | | ALANINE RACEMASE WITH BOUND PROPIONATE INHIBITOR | | Descriptor: | PROPANOIC ACID, PROTEIN (ALANINE RACEMASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Morollo, A.A, Petsko, G.A, Ringe, D. | | Deposit date: | 1999-02-16 | | Release date: | 1999-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Michaelis complex analogue: propionate binds in the substrate carboxylate site of alanine racemase.
Biochemistry, 38, 1999
|
|
1B9Y
 
 | |
7MDH
 
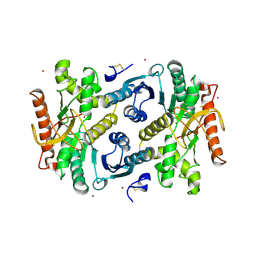 | | STRUCTURAL BASIS FOR LIGHT ACITVATION OF A CHLOROPLAST ENZYME. THE STRUCTURE OF SORGHUM NADP-MALATE DEHYDROGENASE IN ITS OXIDIZED FORM | | Descriptor: | PROTEIN (MALATE DEHYDROGENASE), ZINC ION | | Authors: | Johansson, K, Ramaswamy, S, Saarinen, M, Lemaire-Chamley, M, Issakidis-Bourguet, E, Miginiac-Maslow, M, Eklund, H. | | Deposit date: | 1999-02-16 | | Release date: | 1999-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for light activation of a chloroplast enzyme: the structure of sorghum NADP-malate dehydrogenase in its oxidized form.
Biochemistry, 38, 1999
|
|
1B9X
 
 | |
1XBH
 
 | | A BETA-HAIRPIN MIMIC FROM FCERI-ALPHA-CYCLO(L-262) | | Descriptor: | PROTEIN (CYCLO(L-262)) | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Sutton, B.J, Cowburn, D. | | Deposit date: | 1999-02-17 | | Release date: | 1999-02-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of FceRI Alpha-Chain Mimics: A Beta-Hairpin Peptide and Its Retroenantiomer
J.Am.Chem.Soc., 119, 1997
|
|
2GPA
 
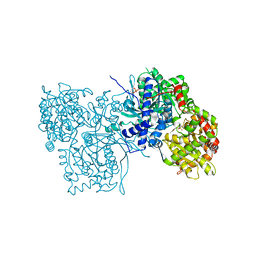 | | ALLOSTERIC INHIBITION OF GLYCOGEN PHOSPHORYLASE A BY A POTENTIAL ANTIDIABETIC DRUG | | Descriptor: | GLYCEROL, PROTEIN (GLYCOGEN PHOSPHORYLASE), PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Oikonomakos, N.G, Tsitsanou, K.E, Zographos, S.E, Skamnaki, V.T. | | Deposit date: | 1999-02-18 | | Release date: | 1999-02-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric inhibition of glycogen phosphorylase a by the potential antidiabetic drug 3-isopropyl 4-(2-chlorophenyl)-1,4-dihydro-1-ethyl-2-methyl-pyridine-3,5,6-tricarbo xylate.
Protein Sci., 8, 1999
|
|
2BVW
 
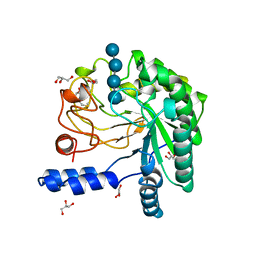 | | CELLOBIOHYDROLASE II (CEL6A) FROM HUMICOLA INSOLENS IN COMPLEX WITH GLUCOSE AND CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE II, GLYCEROL, ... | | Authors: | Varrot, A, Davies, G.J, Schulein, M. | | Deposit date: | 1999-02-18 | | Release date: | 2000-02-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural changes of the active site tunnel of Humicola insolens cellobiohydrolase, Cel6A, upon oligosaccharide binding.
Biochemistry, 38, 1999
|
|
453D
 
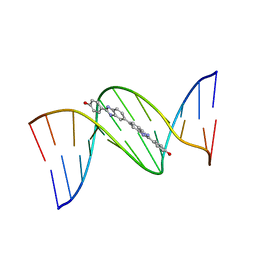 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'-BENZIMIDAZOLE COMPLEX | | Descriptor: | 4-{[4-HYDROXY-PHENYL]-1H-BENZIMIDAZOLE-5-YL}-BENZIMIDAZOLE-2-YL-[4-HYDROXY-BENZENE], DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Neidle, S. | | Deposit date: | 1999-02-18 | | Release date: | 2000-05-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Symmetric bis-benzimidazoles, a new class of sequence-selective DNA-binding molecules
J.Chem.Soc.,Chem.Commun., 1999
|
|
452D
 
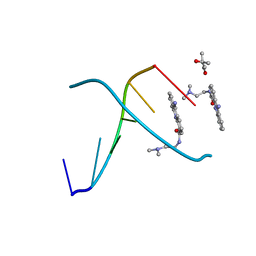 | | ACRIDINE BINDING TO DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Thorpe, J.H, Todd, A.K, Cardin, C.J. | | Deposit date: | 1999-02-18 | | Release date: | 2003-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Major groove binding and 'DNA-induced' fit in the intercalation of a derivative of the mixed topoisomerase I/II poison N-(2-(dimethlyamino)ethyl)acridine-4-carboxamide (DACA) into DNA: X-ray structure complexed to d(CG(5Br-U)ACG)2 at 1.3-angstrom resolution
J.Med.Chem., 42, 1999
|
|
2CUA
 
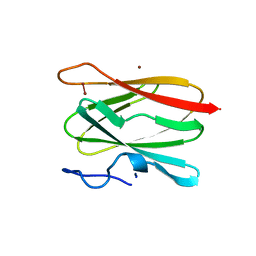 | | THE CUA DOMAIN OF CYTOCHROME BA3 FROM THERMUS THERMOPHILUS | | Descriptor: | DINUCLEAR COPPER ION, PROTEIN (CUA), ZINC ION | | Authors: | Williams, P.A, Blackburn, N.J, Sanders, D, Bellamy, H, Stura, E.A, Fee, J.A, Mcree, D.E. | | Deposit date: | 1999-02-18 | | Release date: | 1999-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The CuA domain of Thermus thermophilus ba3-type cytochrome c oxidase at 1.6 A resolution.
Nat.Struct.Biol., 6, 1999
|
|
3AMV
 
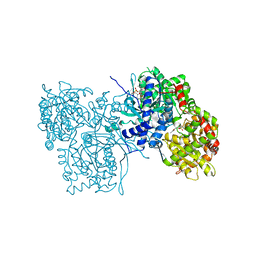 | | ALLOSTERIC INHIBITION OF GLYCOGEN PHOSPHORYLASE A BY A POTENTIAL ANTIDIABETIC DRUG | | Descriptor: | 2,3-DICARBOXY-4-(2-CHLORO-PHENYL)-1-ETHYL-5-ISOPROPOXYCARBONYL-6-METHYL-PYRIDINIUM, GLYCEROL, PROTEIN (GLYCOGEN PHOSPHORYLASE), ... | | Authors: | Oikonomakos, N.G, Tsitsanou, K.E, Zographos, S.E, Skamnaki, V.T. | | Deposit date: | 1999-02-18 | | Release date: | 1999-02-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allosteric inhibition of glycogen phosphorylase a by the potential antidiabetic drug 3-isopropyl 4-(2-chlorophenyl)-1,4-dihydro-1-ethyl-2-methyl-pyridine-3,5,6-tricarbo xylate.
Protein Sci., 8, 1999
|
|
1F7M
 
 | | THE FIRST EGF-LIKE DOMAIN FROM HUMAN BLOOD COAGULATION FVII, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN (Blood Coagulation Factor VII) | | Authors: | Kao, Y.-H, Lee, G.F, Wang, Y, Starovasnik, M.A, Kelley, R.F, Spellman, M.W, Lerner, L. | | Deposit date: | 1999-02-19 | | Release date: | 1999-06-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The effect of O-fucosylation on the first EGF-like domain from human blood coagulation factor VII.
Biochemistry, 38, 1999
|
|
1F7E
 
 | | THE FIRST EGF-LIKE DOMAIN FROM HUMAN BLOOD COAGULATION FVII, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (Blood Coagulation Factor VII) | | Authors: | Kao, Y.-H, Lee, G.F, Wang, Y, Starovasnik, M.A, Kelley, R.F, Spellman, M.W, Lerner, L. | | Deposit date: | 1999-02-19 | | Release date: | 1999-06-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The effect of O-fucosylation on the first EGF-like domain from human blood coagulation factor VII.
Biochemistry, 38, 1999
|
|
1FF7
 
 | | THE FIRST EGF-LIKE DOMAIN FROM HUMAN BLOOD COAGULATION FVII (FUCOSYLATED AT SER-60), NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (Blood Coagulation Factor VII), alpha-L-fucopyranose | | Authors: | Kao, Y.-H, Lee, G.F, Wang, Y, Starovasnik, M.A, Kelley, R.F, Spellman, M.W, Lerner, L. | | Deposit date: | 1999-02-19 | | Release date: | 1999-06-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The effect of O-fucosylation on the first EGF-like domain from human blood coagulation factor VII.
Biochemistry, 38, 1999
|
|
1FFM
 
 | | THE FIRST EGF-LIKE DOMAIN FROM HUMAN BLOOD COAGULATION FVII (FUCOSYLATED AT SER-60), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN (Blood Coagulation Factor VII), alpha-L-fucopyranose | | Authors: | Kao, Y.-H, Lee, G.F, Wang, Y, Starovasnik, M.A, Kelley, R.F, Spellman, M.W, Lerner, L. | | Deposit date: | 1999-02-19 | | Release date: | 1999-06-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The effect of O-fucosylation on the first EGF-like domain from human blood coagulation factor VII.
Biochemistry, 38, 1999
|
|
1B97
 
 | |
